| Sequence |
Count |
Percentage |
Possible Source |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
15953 |
0.15729310331907873 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
15313 |
0.1509828427960291 |
No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG |
13985 |
0.13788905221070116 |
No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG |
13910 |
0.1371495685556563 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
13739 |
0.13546354582215397 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
13686 |
0.13494097737258892 |
No Hit |
| CATGGGGTGGGTGCTTGCAAGTGTAAGCCTCGGCAAACAAGGCCTGGCTT |
13016 |
0.12833492338752137 |
No Hit |
| GGCTGCTTGTTGTACGTGGCCTGGTGGAACGCACTGCAAAACGAGCTCAG |
12933 |
0.12751656147593837 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
12643 |
0.12465722467643155 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
12584 |
0.1240754975344629 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
12573 |
0.12396703993172299 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
12570 |
0.12393746058552119 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
12444 |
0.1226951280450458 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
12418 |
0.12243877371129691 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
11835 |
0.11669052076608141 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
11817 |
0.11651304468887062 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
11709 |
0.115448188225606 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
11532 |
0.11370300679970011 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
11509 |
0.11347623181215301 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
11442 |
0.11281562641364624 |
No Hit |
| CATGTTTGGGCCCCTGTTCCCGGTCTTTGGTAAACAGACGCTTAAGTTGA |
11318 |
0.11159301343730539 |
No Hit |
| AGTGACAAGTTGGGAATGATGTGGTGATTTAGAATGCAGTATTGGCCAAG |
11256 |
0.11098170694913496 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
10963 |
0.10809279080342632 |
No Hit |
| CATGGCTCTGCTGTGCCTTCCATCCTGGGCTCCCTTCTCTCCTGTGACCT |
10936 |
0.10782657668761016 |
No Hit |
| ACAGTGAAAACAGTGACAATGTTCCAGCAAAGCCACCAGACAAAGTTCTT |
10930 |
0.10776741799520656 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
10719 |
0.10568700397901366 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
10576 |
0.10427705514339475 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10550 |
0.10402070080964586 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10534 |
0.10386294429656961 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
10527 |
0.10379392582209876 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
10520 |
0.10372490734762792 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
10474 |
0.10327135737253372 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality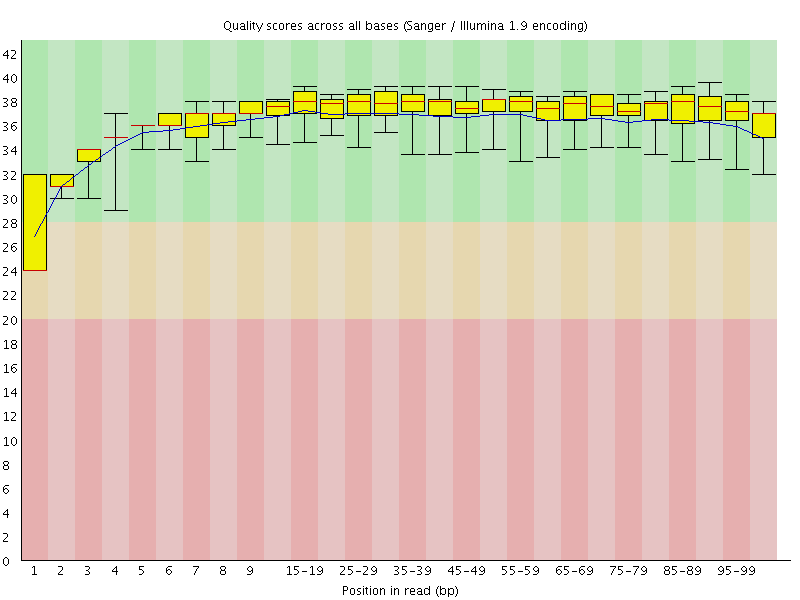
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores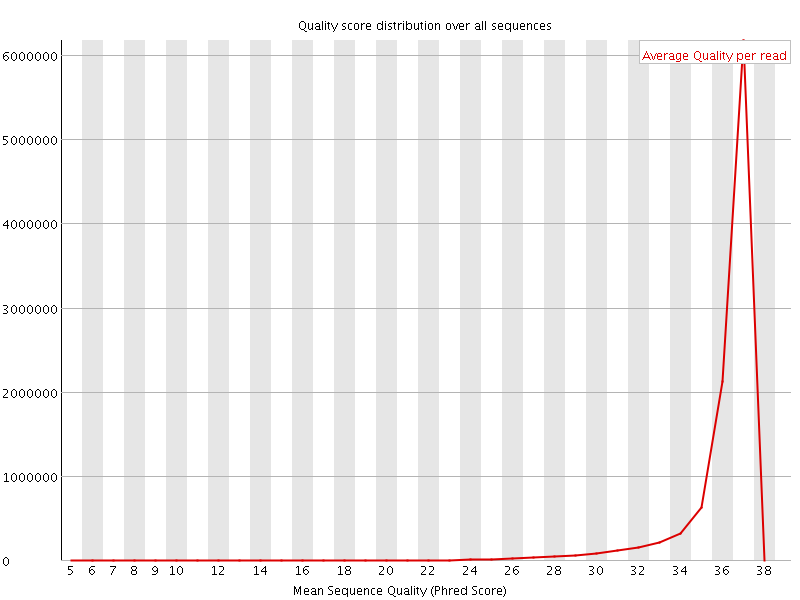
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content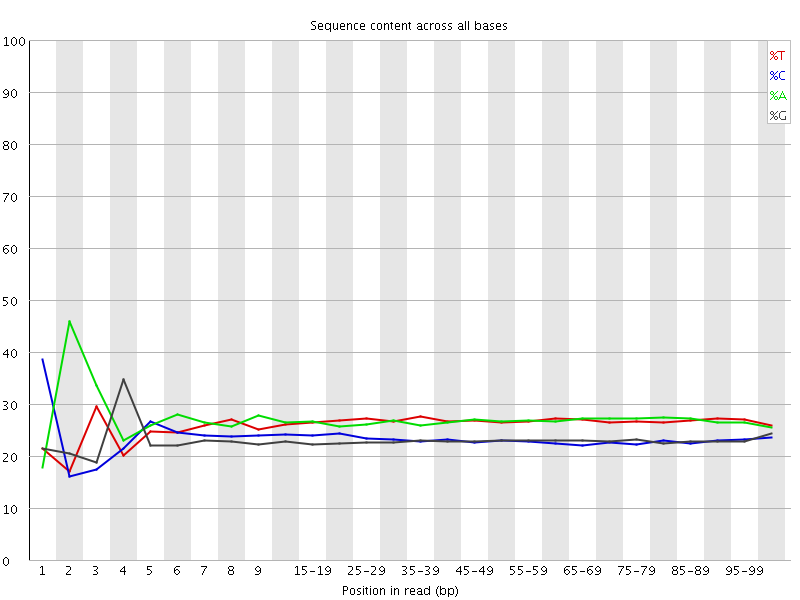
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content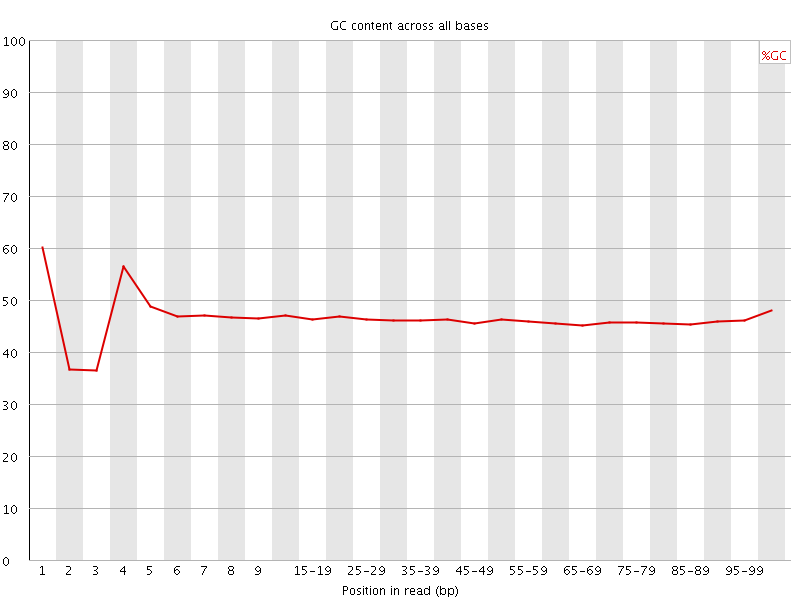
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content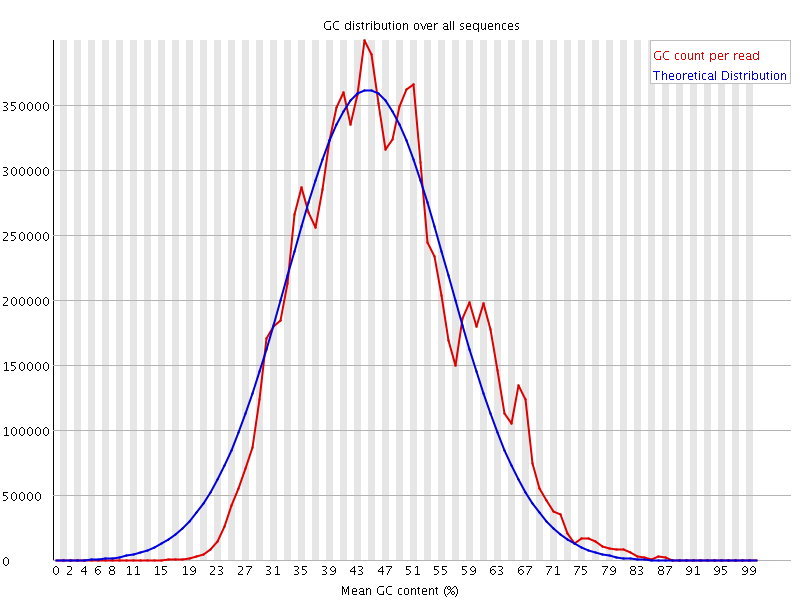
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution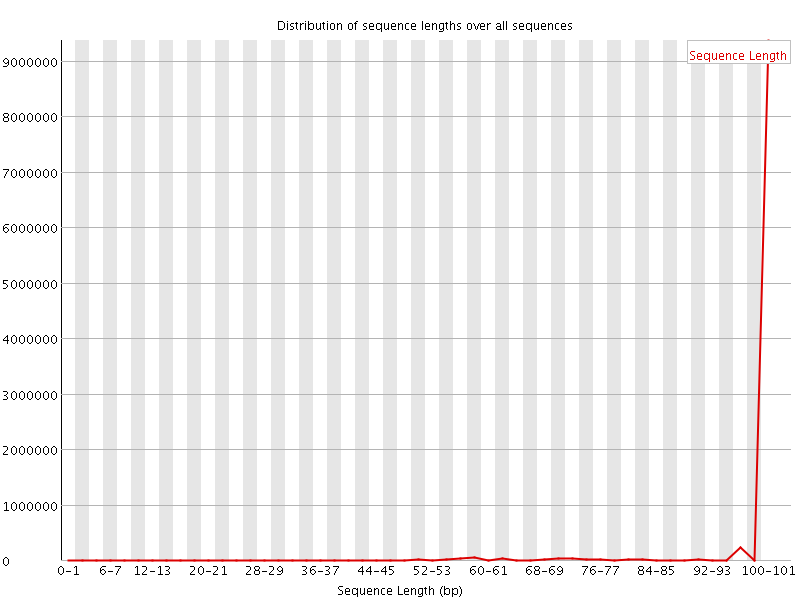
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels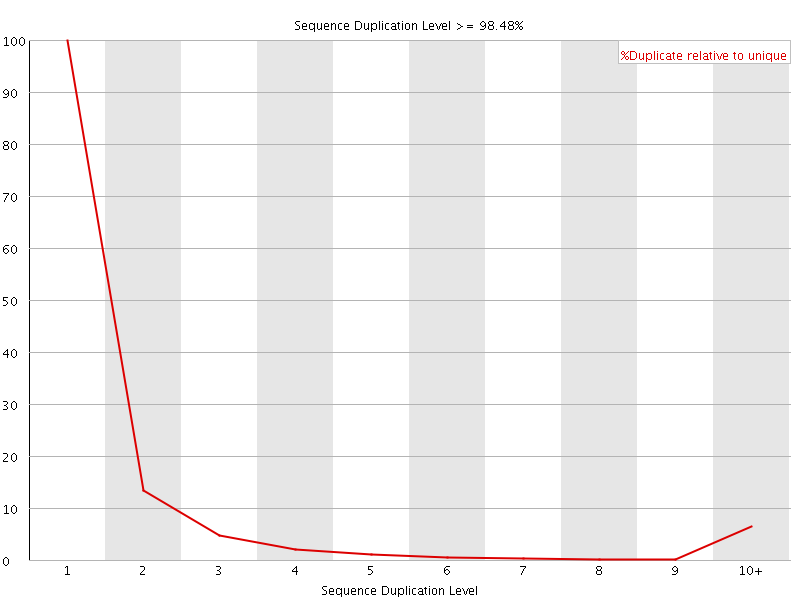
![[WARN]](Icons/warning.png) Overrepresented sequences
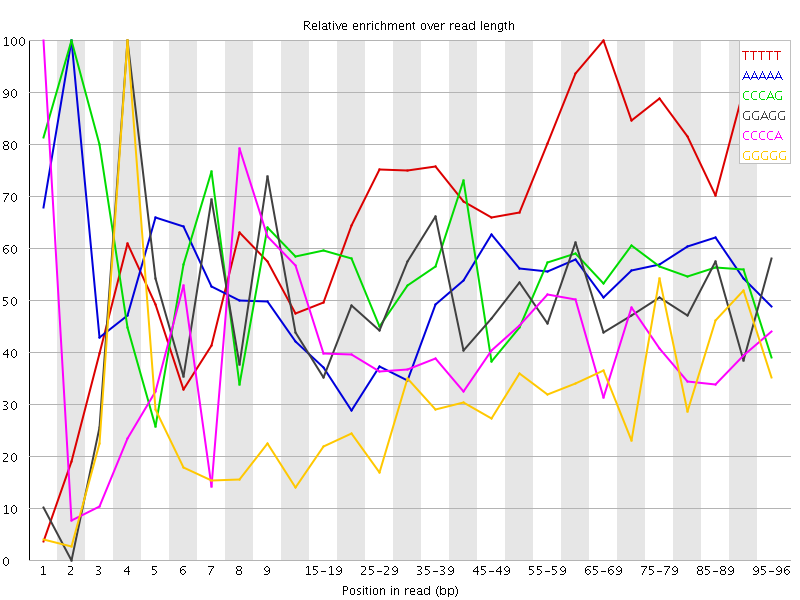
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content