| Sequence |
Count |
Percentage |
Possible Source |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
14726 |
0.16266294461683523 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
14247 |
0.1573719252992022 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
13331 |
0.1472538173765469 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
13282 |
0.14671256487850096 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
12513 |
0.13821821444998364 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
12384 |
0.1367932844041075 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11415 |
0.12608974010601479 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11380 |
0.1257031311788391 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
11280 |
0.12459853424405141 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
11235 |
0.12410146562339697 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
11130 |
0.12294163884186987 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
11077 |
0.1223562024664324 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
10806 |
0.11936274477315775 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
10735 |
0.11857848094945851 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
10621 |
0.11731924044380054 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
10467 |
0.11561816116422749 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
9960 |
0.11001785470485391 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
9915 |
0.10952078608419945 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
9759 |
0.10779761486593065 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
9686 |
0.10699125910353564 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
9487 |
0.10479311120330814 |
No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG |
9465 |
0.10455009987765485 |
No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG |
9388 |
0.10369956023786832 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
9378 |
0.10358910054438955 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
9329 |
0.10304784804634358 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
9310 |
0.10283797462873392 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
9234 |
0.10199848095829528 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
9075 |
0.10024217183198286 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality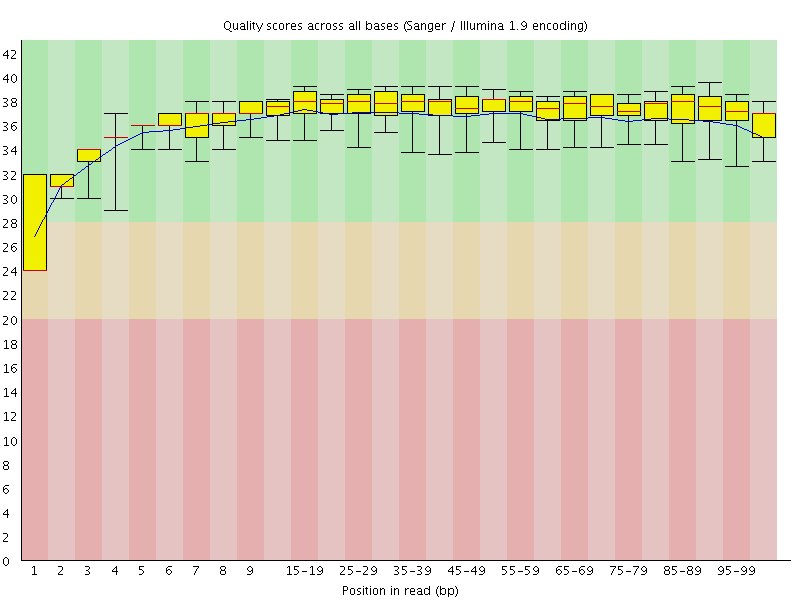
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores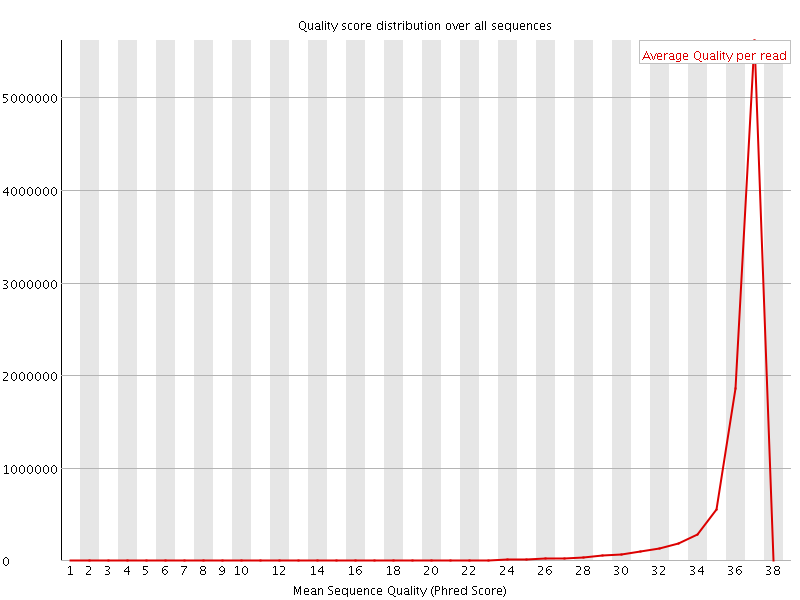
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content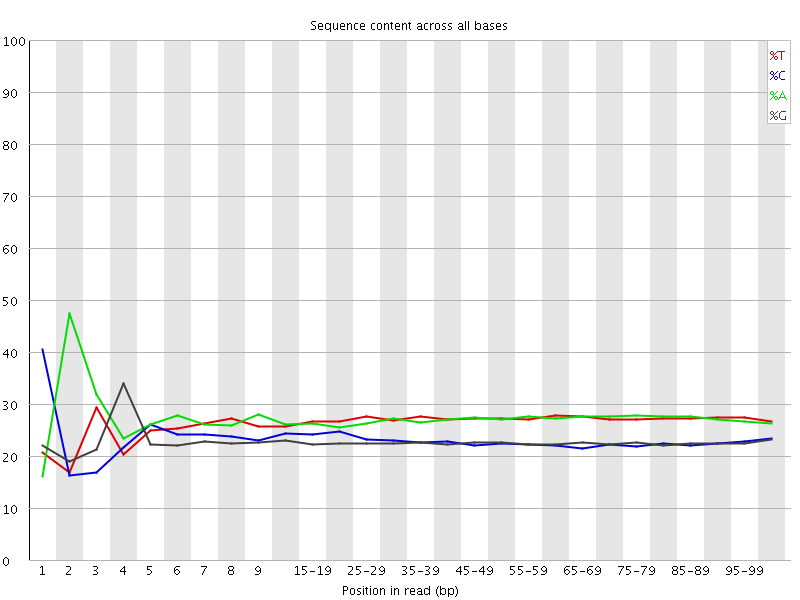
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content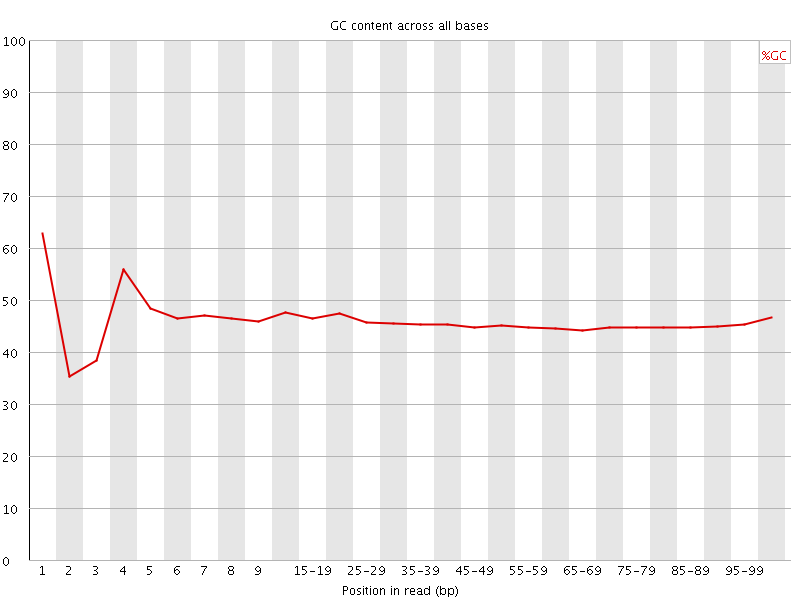
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content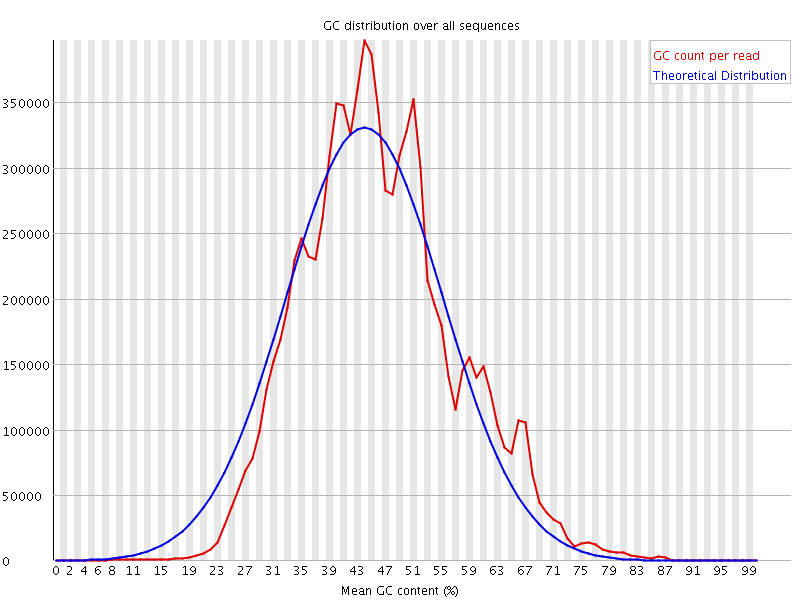
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution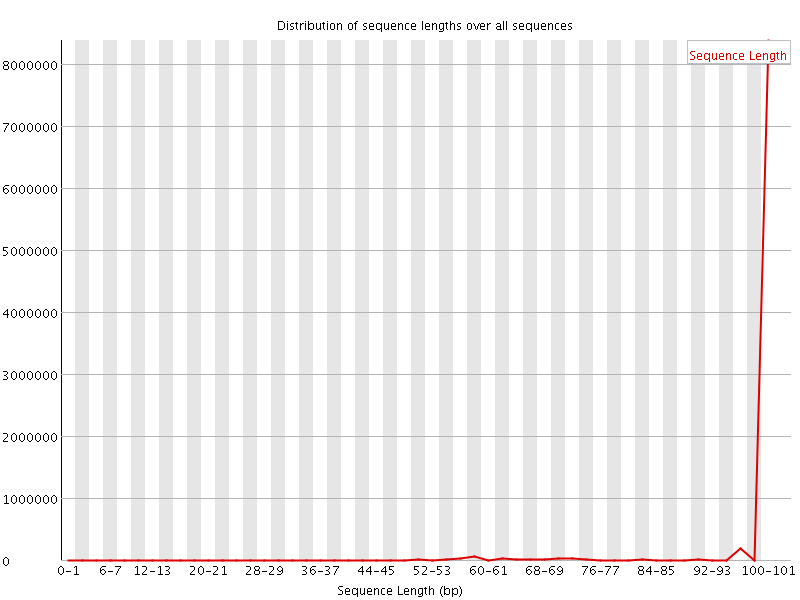
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels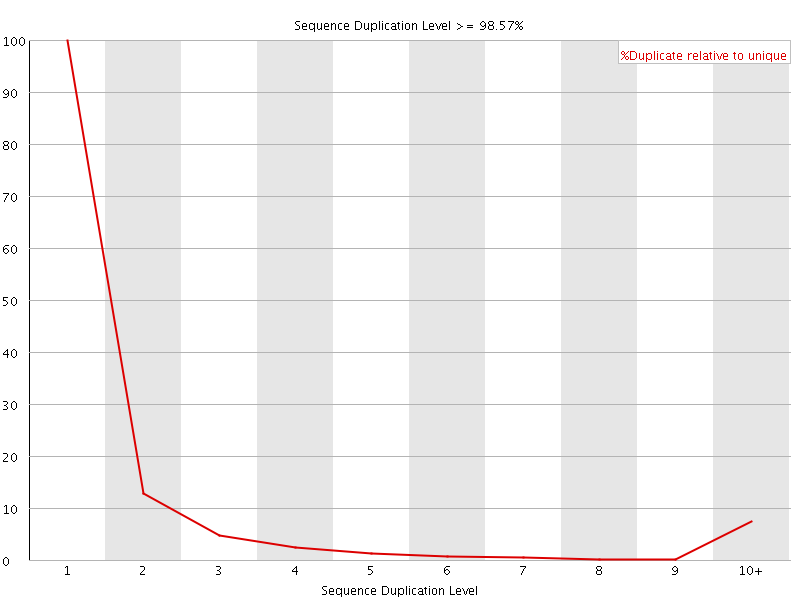
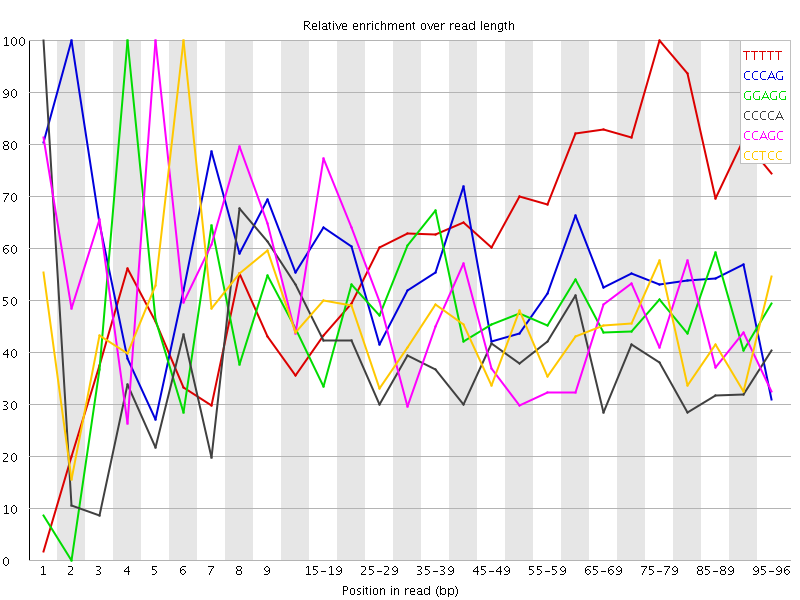
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content