![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-11.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 7538824 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 46 |
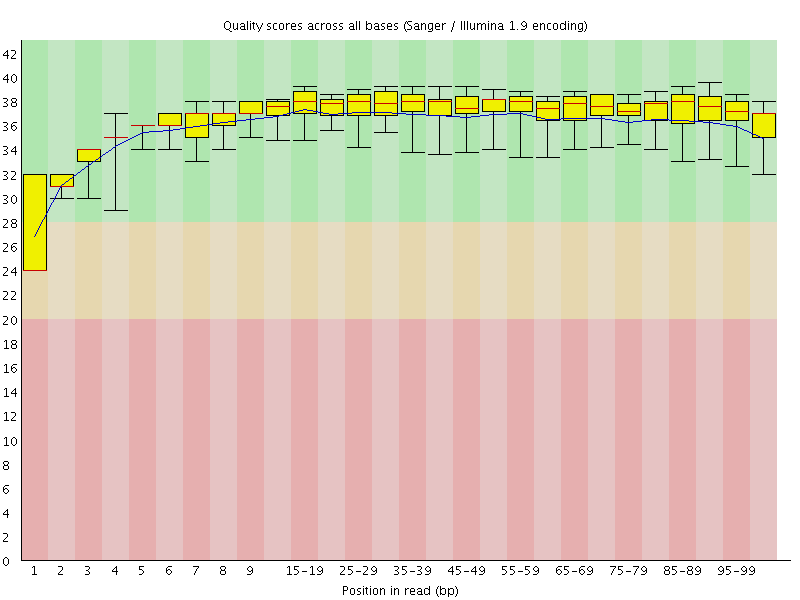
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
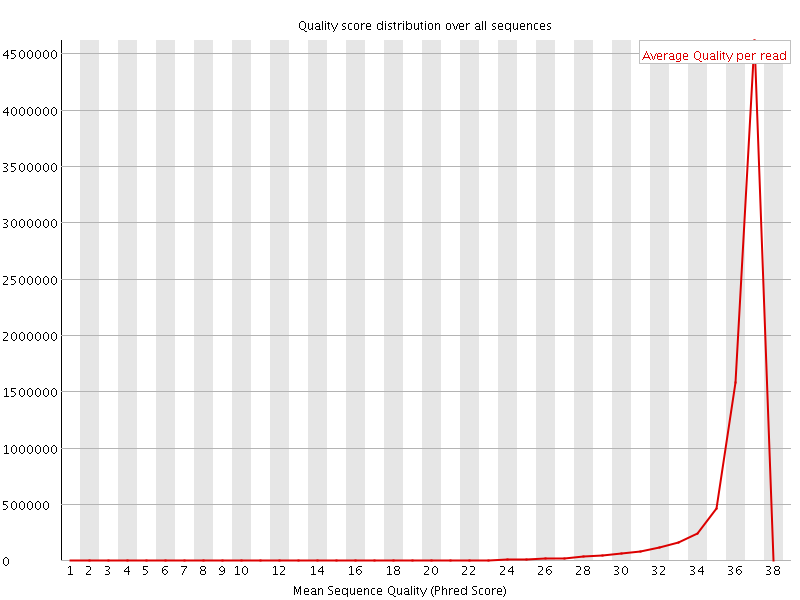
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

![[FAIL]](Icons/error.png) Per base GC content
Per base GC content

![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

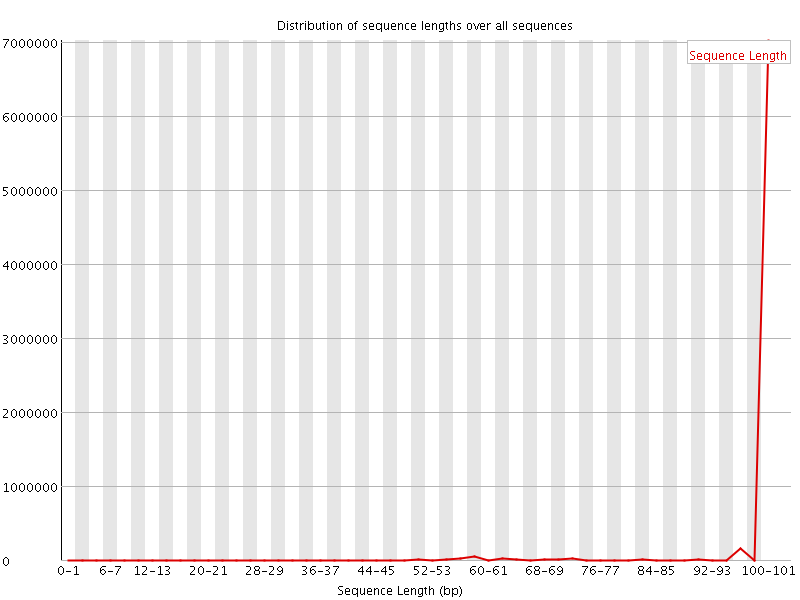
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
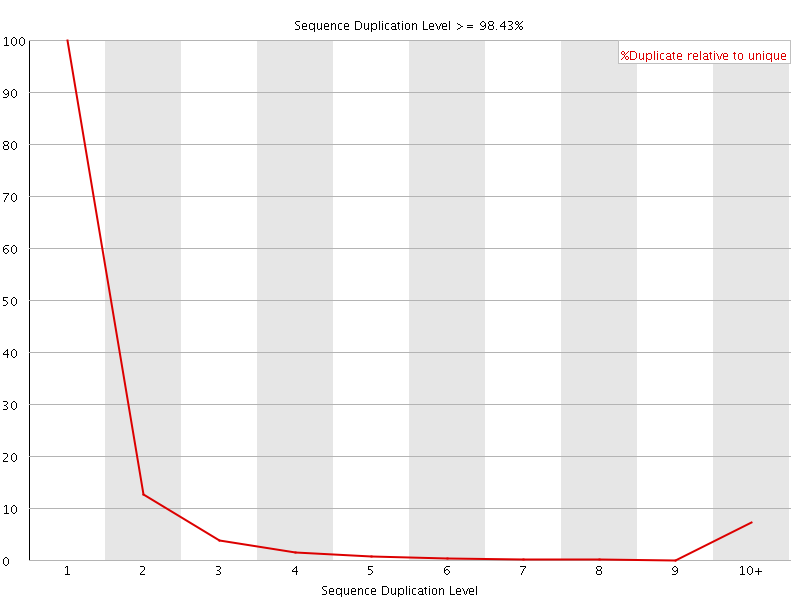
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 11030 | 0.14630929174099302 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 10649 | 0.14125545310515272 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 10202 | 0.1353261463591669 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 10193 | 0.13520676434414705 | No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 9960 | 0.1321160966219665 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 9830 | 0.13039168973834647 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 9129 | 0.12109315723513375 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 9123 | 0.12101356922512051 | No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA | 9104 | 0.12076154052674529 | No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT | 9026 | 0.11972689639657326 | No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT | 9000 | 0.11938201501984924 | No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC | 8963 | 0.11889122229143431 | No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA | 8325 | 0.11042836389336057 | No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA | 8295 | 0.11003042384329438 | No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC | 8276 | 0.10977839514491915 | No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC | 8271 | 0.10971207180324145 | No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT | 7753 | 0.10284097360543235 | No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG | 7731 | 0.10254915090205051 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 7626 | 0.10115636072681894 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 7623 | 0.10111656672181231 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 7612 | 0.1009706553701214 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 7582 | 0.10057271532005521 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
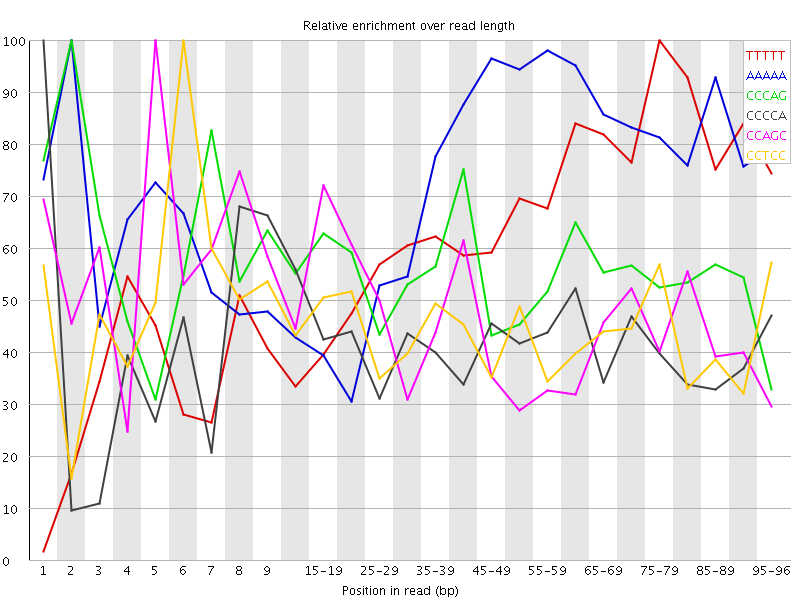
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 3114865 | 3.1677408 | 4.9201937 | 75-79 |
| AAAAA | 3111180 | 3.010305 | 4.102711 | 2 |
| CCCAG | 1605765 | 2.8989747 | 5.1954417 | 2 |
| CCCCA | 1404890 | 2.4989219 | 6.010461 | 1 |
| CCAGC | 1333460 | 2.4073677 | 5.194708 | 5 |
| CCTCC | 1304865 | 2.3442342 | 5.3431873 | 6 |
| TGGGG | 1224630 | 2.3347936 | 8.095843 | 3 |
| CTGCC | 1268920 | 2.3137777 | 5.8767433 | 7 |
| GGGGG | 1019590 | 2.2745867 | 5.6082087 | 4 |
| CCACC | 1191175 | 2.1187804 | 5.9491215 | 1 |
| GGAAG | 1321220 | 2.1102571 | 5.020512 | 4 |
| TGGCT | 1240485 | 1.9913601 | 6.201239 | 3 |
| CCCTG | 1083580 | 1.9758247 | 5.2010345 | 1 |
| TGCAG | 1208810 | 1.9212828 | 5.86736 | 3 |
| GCCCA | 1010890 | 1.8250145 | 5.678838 | 2 |
| TGGCA | 1131560 | 1.7985016 | 5.071086 | 3 |
| GGCTC | 969735 | 1.7947028 | 7.3981256 | 1 |
| GCTCC | 898815 | 1.63892 | 7.159906 | 2 |
| TGCCC | 863560 | 1.5746351 | 9.545005 | 3 |
| TAAAA | 1571640 | 1.5359017 | 5.739783 | 1 |
| TGAGG | 915195 | 1.4763825 | 5.047185 | 1 |
| CATGG | 927510 | 1.4741845 | 36.506924 | 1 |
| ATGGA | 1066210 | 1.4553577 | 10.250294 | 2 |
| GGCCC | 660145 | 1.4085104 | 5.717981 | 1 |
| TGGCC | 733405 | 1.3573235 | 5.213587 | 3 |
| TGGGT | 811820 | 1.3227265 | 5.6585717 | 3 |
| ATGGG | 811225 | 1.3086593 | 11.551987 | 2 |
| ATGTG | 944510 | 1.3021425 | 9.398456 | 2 |
| ATGGC | 812385 | 1.2912047 | 10.995841 | 2 |
| ATGAA | 1024480 | 1.183238 | 7.872505 | 2 |
| ATGCC | 738465 | 1.1564081 | 9.105476 | 2 |
| TGAGC | 710210 | 1.1288078 | 5.112411 | 1 |
| ATGTT | 937790 | 1.1049032 | 6.2615695 | 2 |
| ATGCT | 791615 | 1.0752611 | 8.014821 | 2 |
| CATGA | 797065 | 1.0719355 | 23.255438 | 1 |
| CATGT | 782230 | 1.0625134 | 27.746927 | 1 |
| ATGAG | 755585 | 1.0313599 | 5.4783826 | 2 |
| ATGGT | 721765 | 0.99505657 | 6.295585 | 2 |
| CATGC | 625770 | 0.979932 | 28.781073 | 1 |
| ATGAC | 692920 | 0.9318757 | 6.0510993 | 2 |
| ATGTA | 749035 | 0.87376726 | 7.1057076 | 2 |
| ATGCA | 604460 | 0.81291 | 9.89845 | 2 |
| TAAGT | 640935 | 0.747666 | 6.0705323 | 1 |