| Sequence |
Count |
Percentage |
Possible Source |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
12760 |
0.15931905338511784 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
12758 |
0.1592940817466562 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
12707 |
0.15865730496588498 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
12522 |
0.15634742840818538 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
12419 |
0.15506138902741212 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
11972 |
0.14948022783124068 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11924 |
0.14888090850816185 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11920 |
0.14883096523123862 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
11538 |
0.14406138228506973 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
11333 |
0.14150178934275395 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
10808 |
0.13494673424657944 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
10787 |
0.13468453204273245 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
9949 |
0.12422141552731485 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
9879 |
0.12334740818115825 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
9762 |
0.12188656733115365 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
9681 |
0.12087521597345814 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
9617 |
0.1200761235426864 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
9614 |
0.12003866608499397 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
9253 |
0.11553128534267207 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
9240 |
0.11536896969267155 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
8985 |
0.11218508578881535 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
8970 |
0.11199779850035321 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
8780 |
0.1096254928464996 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
8751 |
0.10926340408880615 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
8750 |
0.10925091826957532 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
8729 |
0.10898871606572835 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
8583 |
0.1071657864580303 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
8554 |
0.10680369770033685 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
8543 |
0.10666635368879794 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
8526 |
0.1064540947618742 |
No Hit |
| GACCCCCTCCACTAACTCCCGAGGACGTTGGCTTTGCATCTGGTTTTTCT |
8468 |
0.1057299172464873 |
No Hit |
| AGAAGAGGGCTCCTTTTGTGCATATCAAACCTTGTTCCAGAATGGAGTGG |
8464 |
0.10567997396956406 |
No Hit |
| GATCCAAAACTTCTCTCCGACACTGAATACCTTCTGCTTCTCCTATCATT |
8421 |
0.10514308374263928 |
No Hit |
| CATGTGCCTTCACTGTGTCCCAGGAAATCTGGGTTGGTTCCAGTGGGAAA |
8103 |
0.10117259322724216 |
No Hit |
| CGGGGCTTGCTCTCCCCATCCCTATTTCCCCATCTCCTACTCTTTTGCTA |
8048 |
0.1004858731695477 |
No Hit |
| CATGGCTCTGCTGTGCCTTCCATCCTGGGCTCCCTTCTCTCCTGTGACCT |
8033 |
0.10029858588108556 |
No Hit |
| ACAGTGAAAACAGTGACAATGTTCCAGCAAAGCCACCAGACAAAGTTCTT |
8024 |
0.10018621350800828 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality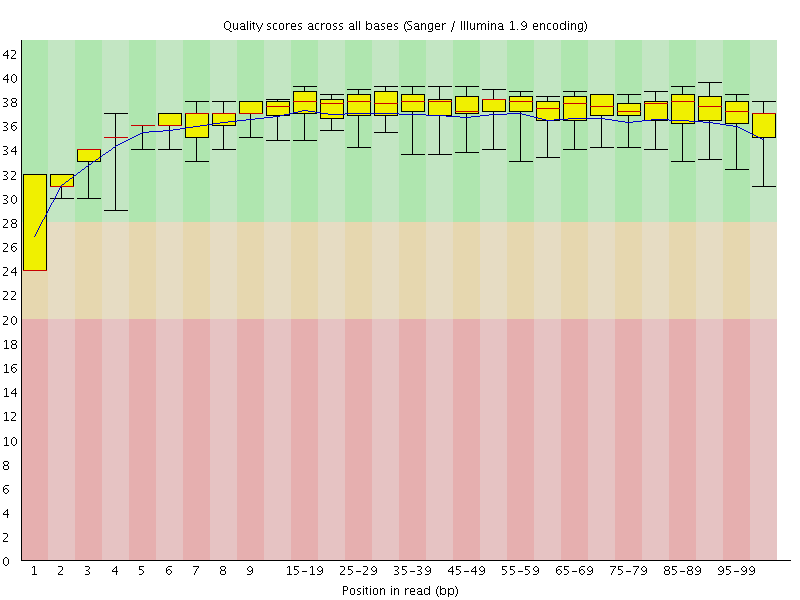
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores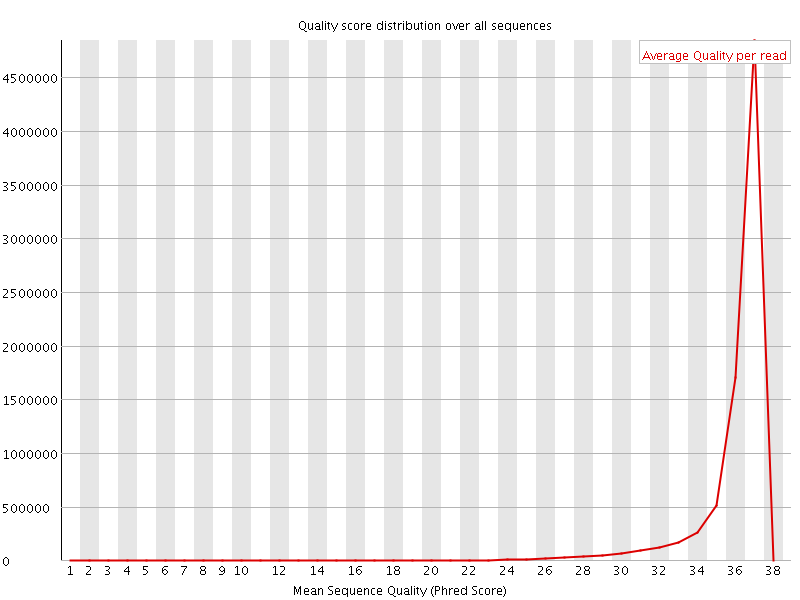
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content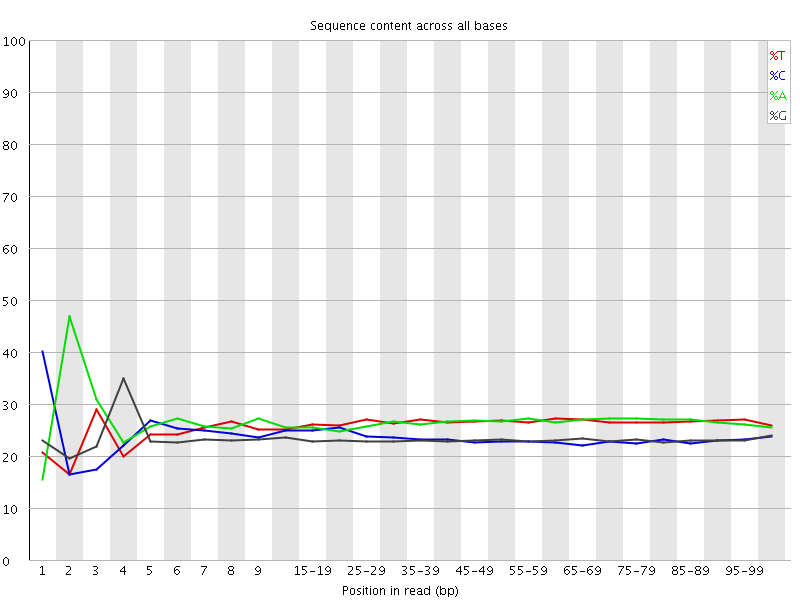
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content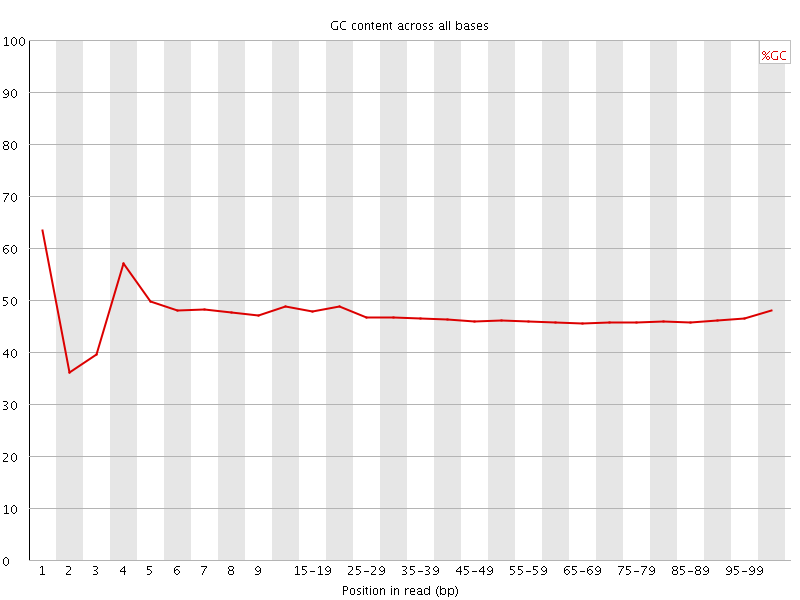
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content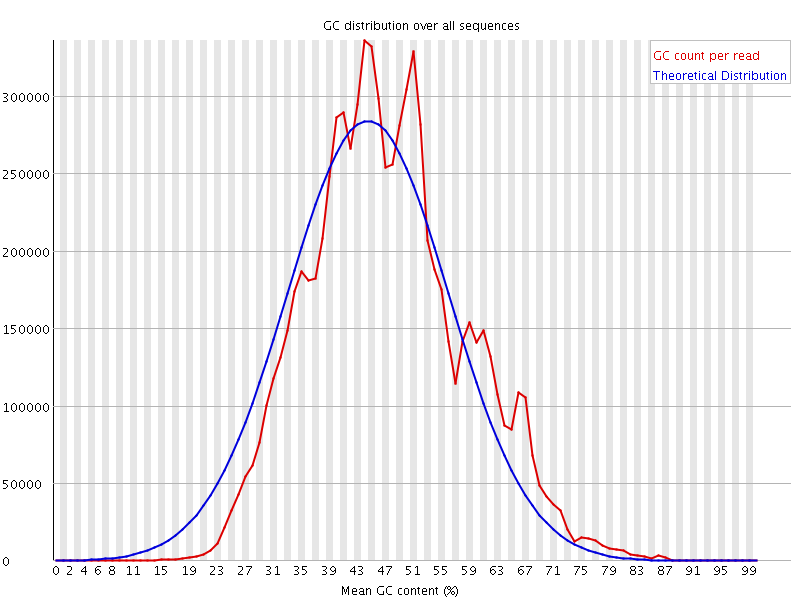
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution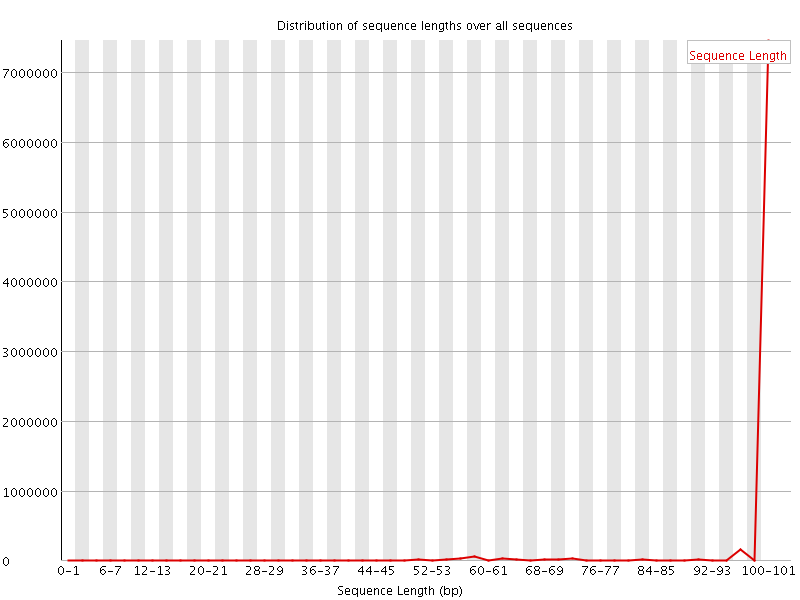
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels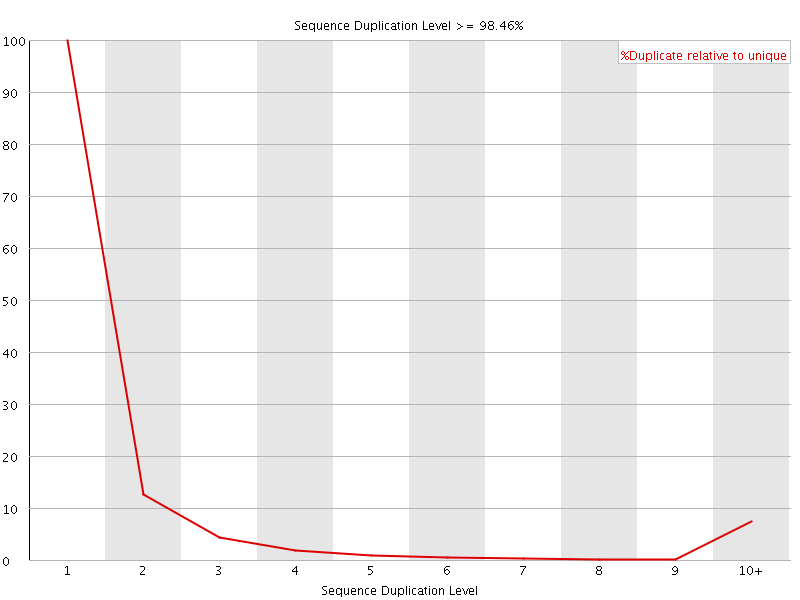
![[WARN]](Icons/warning.png) Overrepresented sequences
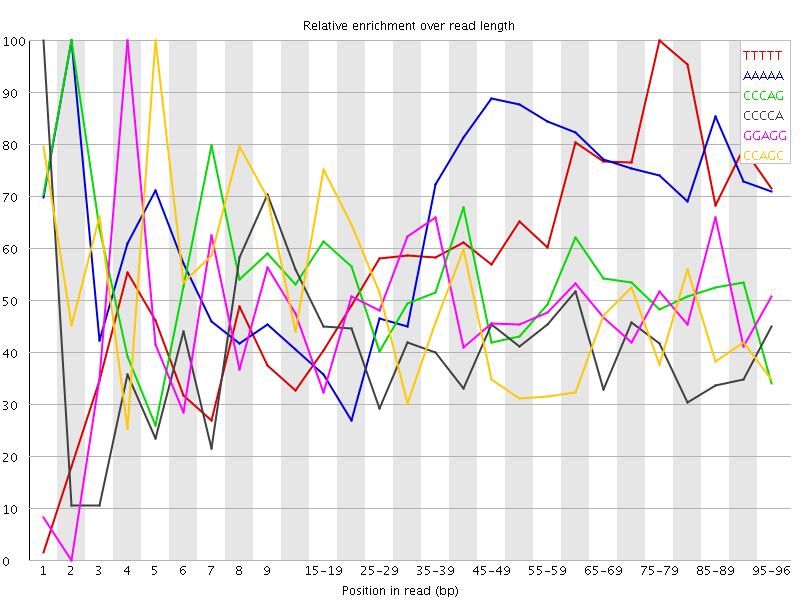
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content