![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-14.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 7507752 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 46 |
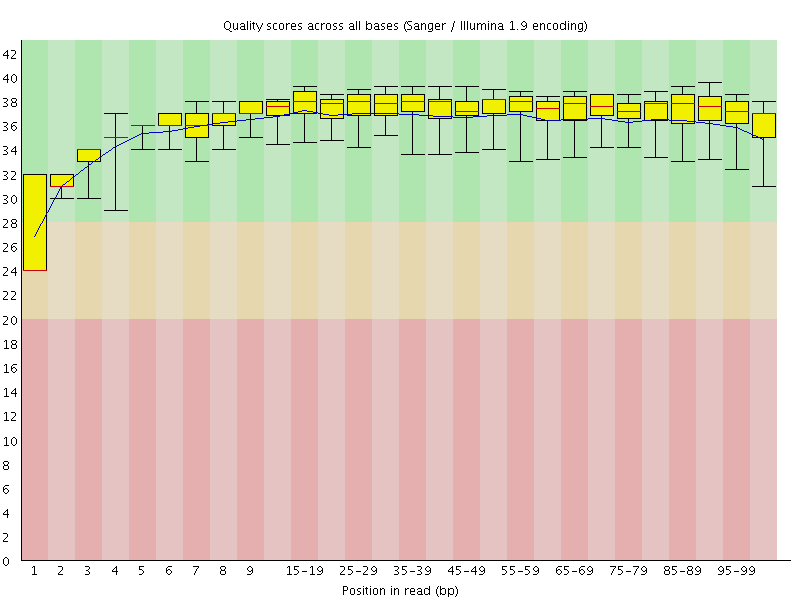
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
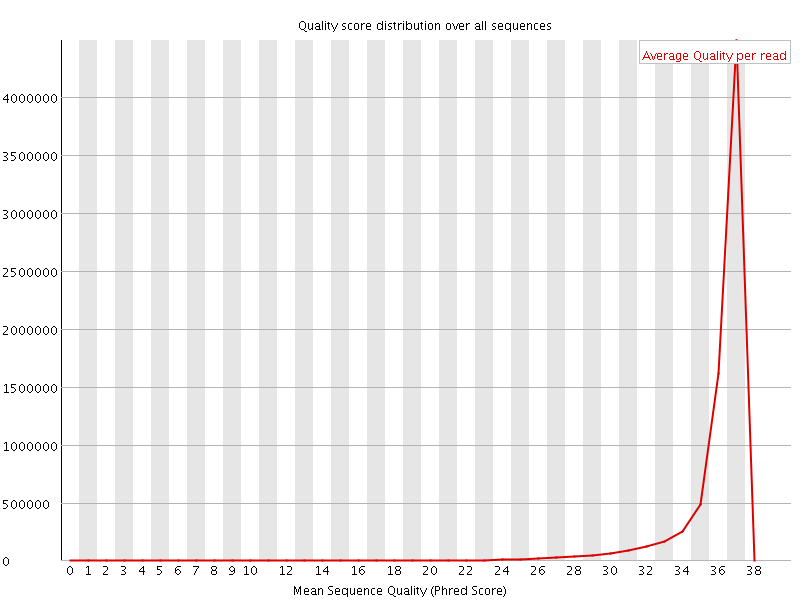
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
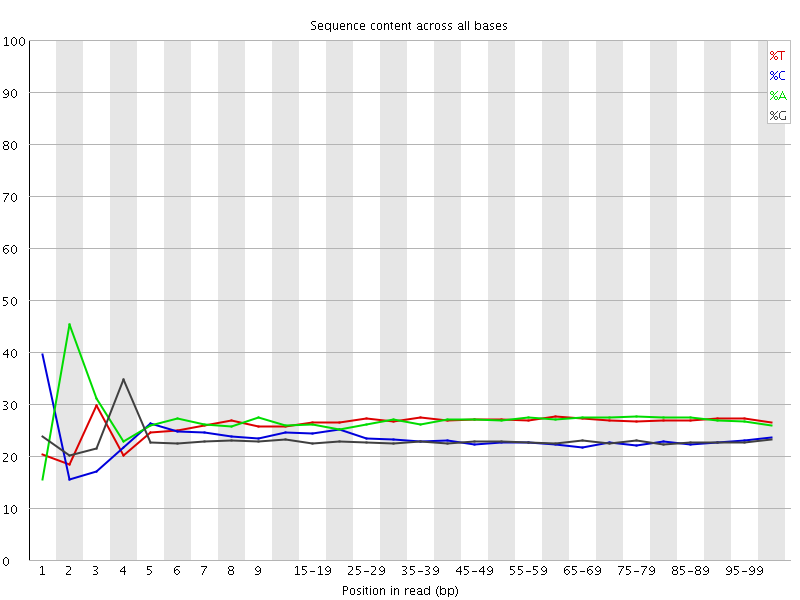
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
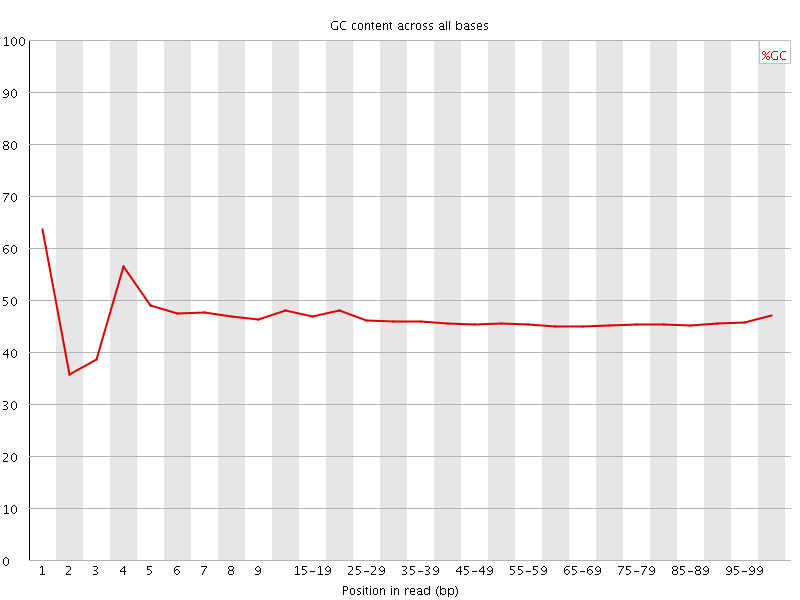
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
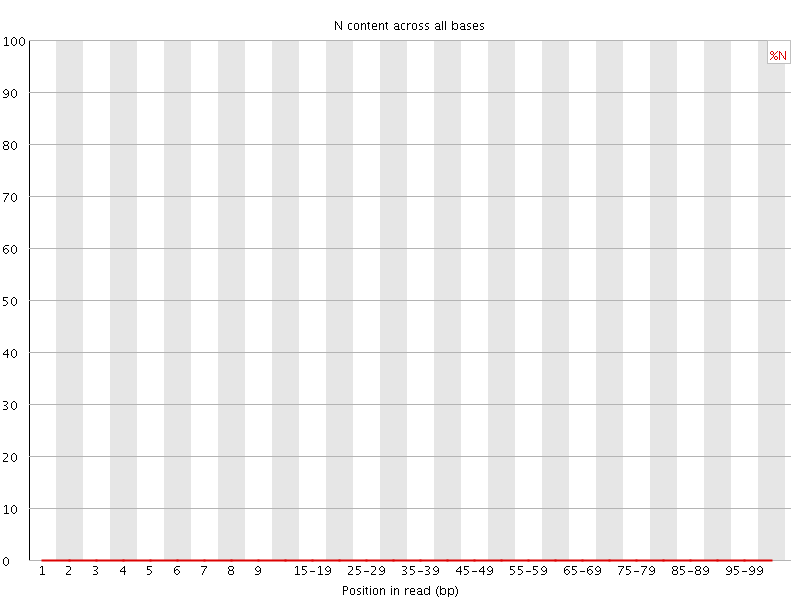
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
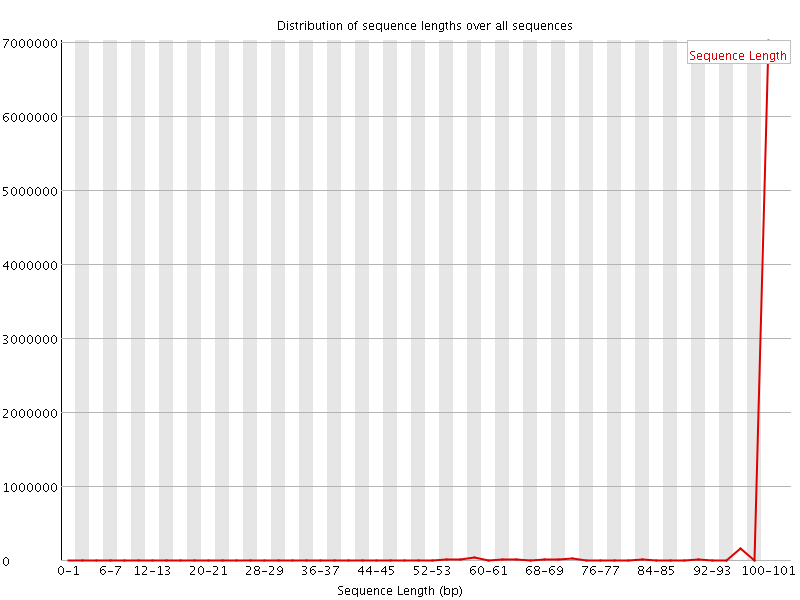
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 10347 | 0.13781755177848176 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 10237 | 0.13635239949321715 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 9250 | 0.12320598762452463 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 9236 | 0.12301951369730912 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 8646 | 0.11516096962179892 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 8639 | 0.11506773265819116 | No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 8610 | 0.11468146523753049 | No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT | 8387 | 0.11171120196831223 | No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT | 8354 | 0.11127165628273283 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 8296 | 0.1104991214414115 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 8169 | 0.10880753653024235 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 8143 | 0.10846122780827071 | No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT | 8075 | 0.10755549730465258 | No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG | 7980 | 0.10629013851283313 | No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA | 7873 | 0.10486494492625756 | No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC | 7851 | 0.10457191446920464 | No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA | 7718 | 0.10280041216065741 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 7666 | 0.10210779471671413 | No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC | 7635 | 0.10169488816359411 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 7634 | 0.10168156859736444 | No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA | 7626 | 0.101575012067527 | No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC | 7595 | 0.10116210551440698 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
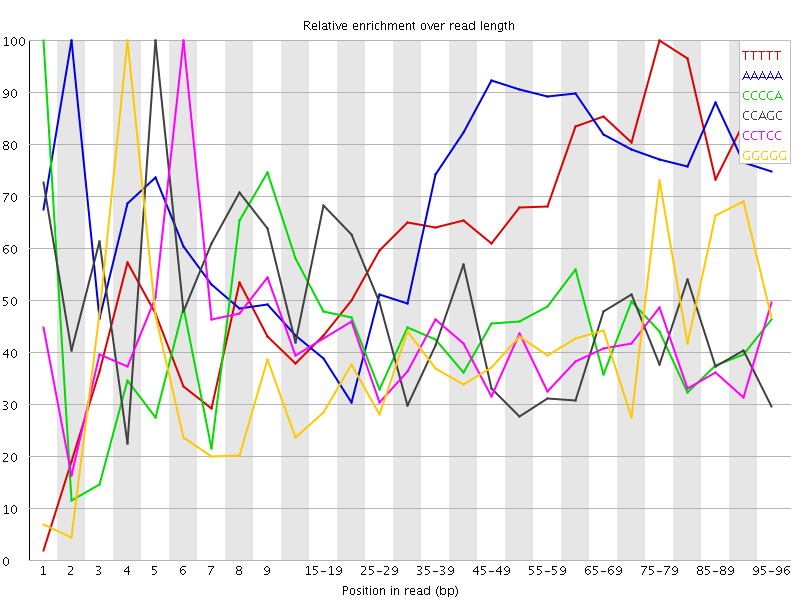
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 3103535 | 3.1576824 | 4.746329 | 75-79 |
| AAAAA | 3071085 | 3.0091963 | 4.271723 | 2 |
| CCCCA | 1403880 | 2.51042 | 5.703453 | 1 |
| CCAGC | 1346990 | 2.4401426 | 5.415353 | 5 |
| CCTCC | 1316265 | 2.3715396 | 5.9131656 | 6 |
| GGGGG | 1061700 | 2.3545332 | 5.67865 | 4 |
| TGGGG | 1232205 | 2.3383403 | 7.8389425 | 3 |
| CTGCC | 1253565 | 2.2880647 | 5.2895684 | 7 |
| CCACC | 1173215 | 2.0979447 | 5.4835362 | 1 |
| GGAAG | 1295340 | 2.0719967 | 5.047251 | 4 |
| CCCTG | 1088030 | 1.9859227 | 5.0523577 | 1 |
| TGGCT | 1225155 | 1.9638264 | 6.235578 | 3 |
| TGCAG | 1222565 | 1.9449722 | 5.951864 | 3 |
| GCCCA | 1001435 | 1.8141519 | 5.622545 | 2 |
| GGCTC | 969505 | 1.7926931 | 7.171089 | 1 |
| GCTCC | 894325 | 1.6323633 | 6.848286 | 2 |
| TGCCC | 861490 | 1.5724313 | 9.325171 | 3 |
| TAAAA | 1588125 | 1.5678841 | 5.9693956 | 1 |
| TGAGG | 931905 | 1.5019226 | 5.0039477 | 1 |
| GGCCC | 696765 | 1.4862301 | 5.9447117 | 1 |
| CATGG | 929180 | 1.4782275 | 36.672607 | 1 |
| ATGGA | 1067405 | 1.4610204 | 10.108017 | 2 |
| TGGCC | 742855 | 1.3735989 | 5.103761 | 3 |
| TGGGT | 823555 | 1.3373317 | 5.8617315 | 3 |
| ATGTG | 948790 | 1.3084819 | 9.98032 | 2 |
| ATGGG | 797715 | 1.2856526 | 11.691531 | 2 |
| ATGGC | 792340 | 1.2605296 | 10.640399 | 2 |
| TGAGA | 898350 | 1.2296247 | 5.2908287 | 1 |
| ATGAA | 1032175 | 1.1998625 | 8.318507 | 2 |
| ATGCC | 737570 | 1.1582712 | 8.811735 | 2 |
| TGAGC | 705790 | 1.1228377 | 5.115534 | 1 |
| ATGTT | 930175 | 1.097701 | 6.465527 | 2 |
| CATGA | 805175 | 1.0878845 | 24.03675 | 1 |
| CATGT | 782290 | 1.0649544 | 27.775463 | 1 |
| ATGCT | 773785 | 1.0533762 | 8.322267 | 2 |
| ATGAG | 766605 | 1.0492976 | 5.487634 | 2 |
| ATGGT | 721595 | 0.995156 | 6.63181 | 2 |
| CATGC | 616600 | 0.96830136 | 28.487347 | 1 |
| ATGAC | 697185 | 0.9419777 | 6.508712 | 2 |
| ATGTA | 735335 | 0.86125946 | 6.5233502 | 2 |
| ATGCA | 612010 | 0.8268964 | 9.515828 | 2 |
| TAAGT | 632750 | 0.74110705 | 6.038148 | 1 |