| Sequence |
Count |
Percentage |
Possible Source |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
11740 |
0.1579386928810816 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
11634 |
0.15651267061145682 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
11626 |
0.156405046289221 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
11624 |
0.15637814020866203 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
11582 |
0.15581311251692392 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
11172 |
0.1502973660023376 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
10983 |
0.1477547413895161 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
10931 |
0.1470551832949832 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
10685 |
0.14374573538623142 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
10465 |
0.14078606652474607 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
10073 |
0.13551247473519035 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
10007 |
0.13462457407674475 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
9608 |
0.12925681100523267 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
9563 |
0.12865142419265616 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
9106 |
0.12250338478493432 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
9095 |
0.12235540134186004 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
8956 |
0.12048542874301249 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
8901 |
0.11974551152764115 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
8679 |
0.11675893658559684 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
8678 |
0.11674548354531737 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
8328 |
0.11203691944749977 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
8308 |
0.1117678586419102 |
No Hit |
| CATGTGCCTTCACTGTGTCCCAGGAAATCTGGGTTGGTTCCAGTGGGAAA |
8234 |
0.11077233366122877 |
No Hit |
| CGGGGCTTGCTCTCCCCATCCCTATTTCCCCATCTCCTACTCTTTTGCTA |
8166 |
0.1098575269222242 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
8100 |
0.10896962626377861 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
8092 |
0.10886200194154277 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
7918 |
0.10652117293291345 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
7878 |
0.1059830513217343 |
No Hit |
| CATGGCTCTGCTGTGCCTTCCATCCTGGGCTCCCTTCTCTCCTGTGACCT |
7836 |
0.10541802362999618 |
No Hit |
| ACAGTGAAAACAGTGACAATGTTCCAGCAAAGCCACCAGACAAAGTTCTT |
7827 |
0.10529694626748087 |
No Hit |
| TAGCTAAAAGGGGAAGAAGAGGATCAGCCCAAGGAGGAGGAAGAGGAAAA |
7747 |
0.10422070304512258 |
No Hit |
| GCTCCGCAGGGATCGGGACACTGGACACACGTTCCTCTCCTCTGCACTGG |
7746 |
0.10420725000484309 |
No Hit |
| CATGGGGTGGGTGCTTGCAAGTGTAAGCCTCGGCAAACAAGGCCTGGCTT |
7661 |
0.10306374158108739 |
No Hit |
| GGCTGCTTGTTGTACGTGGCCTGGTGGAACGCACTGCAAAACGAGCTCAG |
7569 |
0.10182606187537534 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
7551 |
0.10158390715034472 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
7488 |
0.10073636561273756 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
7451 |
0.10023860312239684 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
7449 |
0.10021169704183788 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality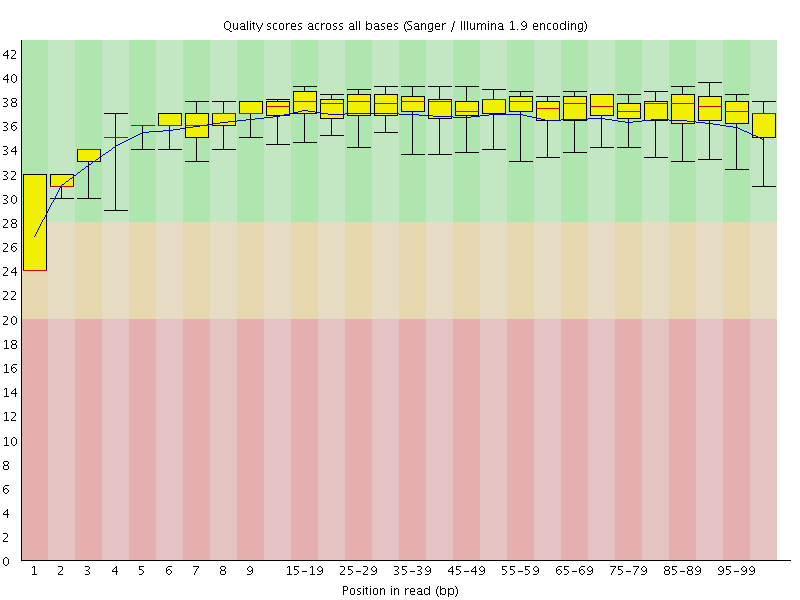
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores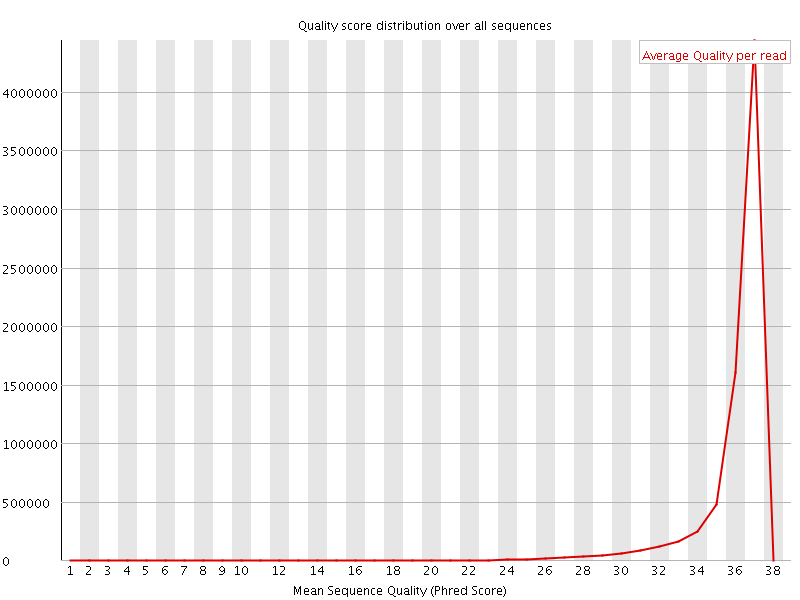
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content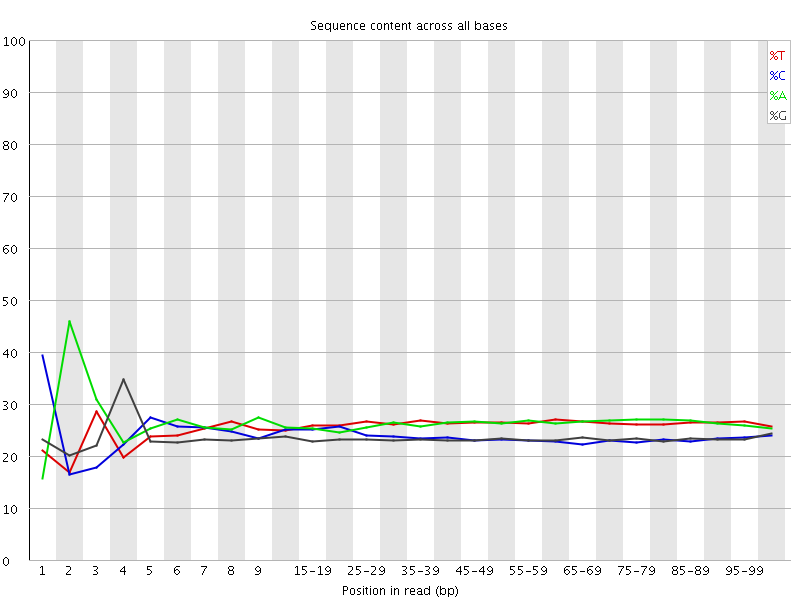
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content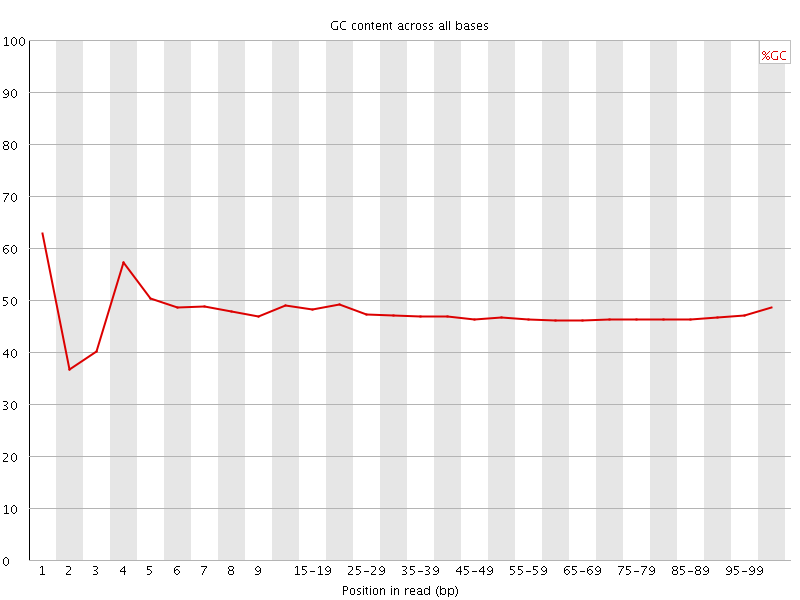
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content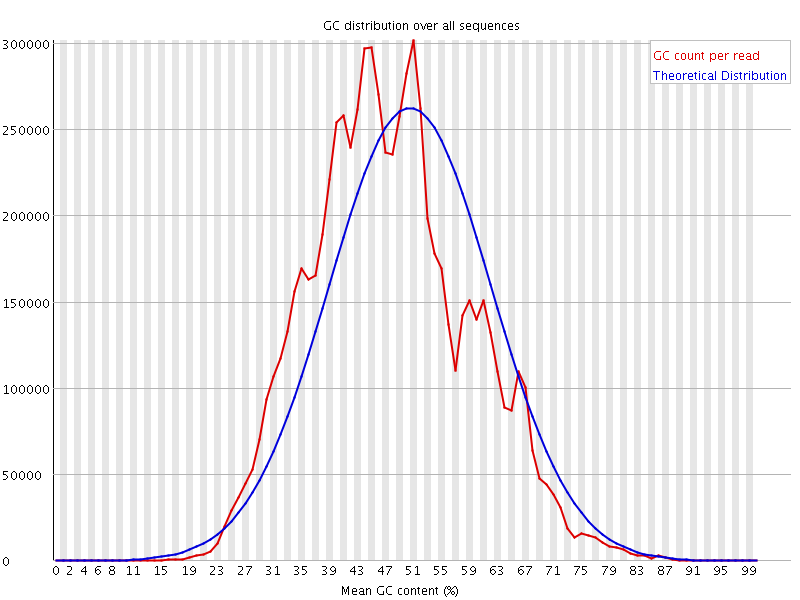
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution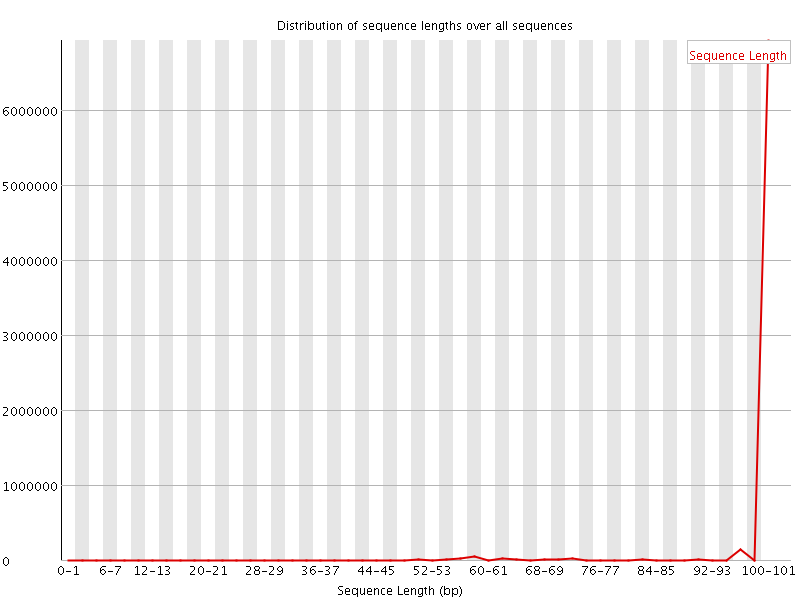
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels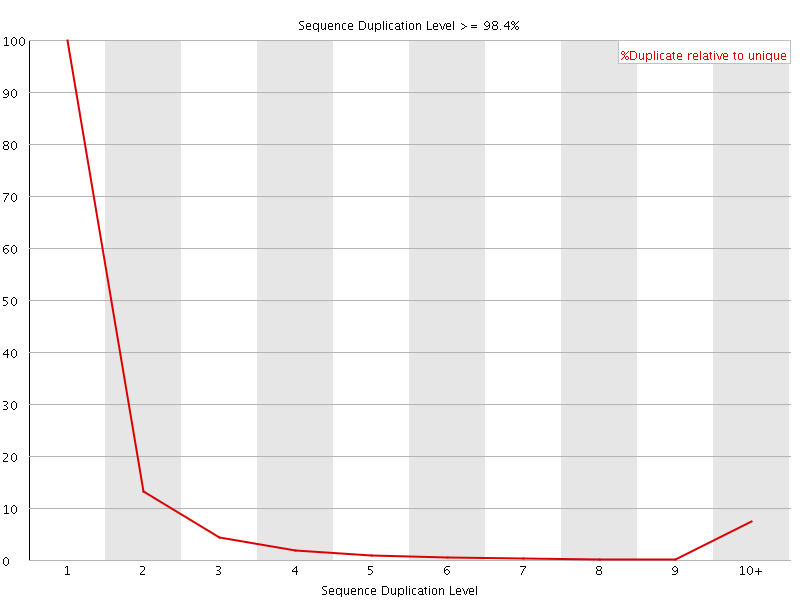
![[WARN]](Icons/warning.png) Overrepresented sequences
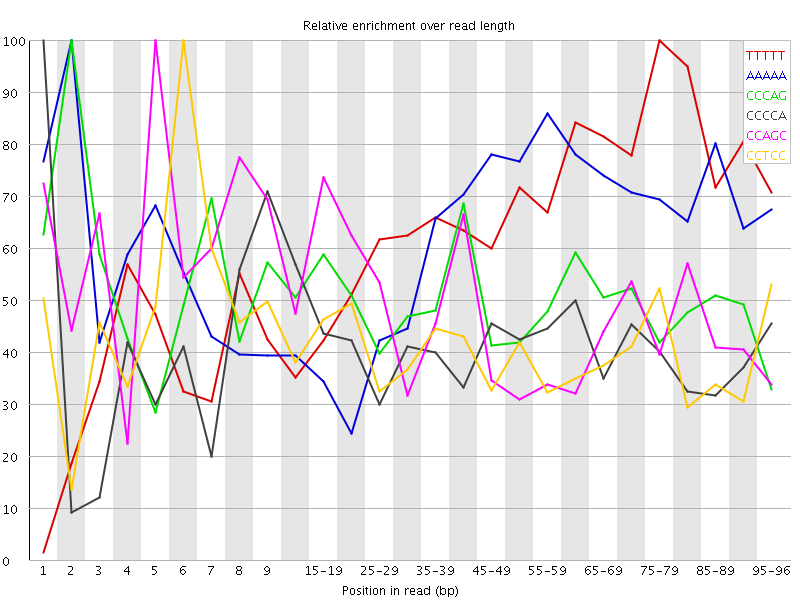
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content