![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-25.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 12017720 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 44 |
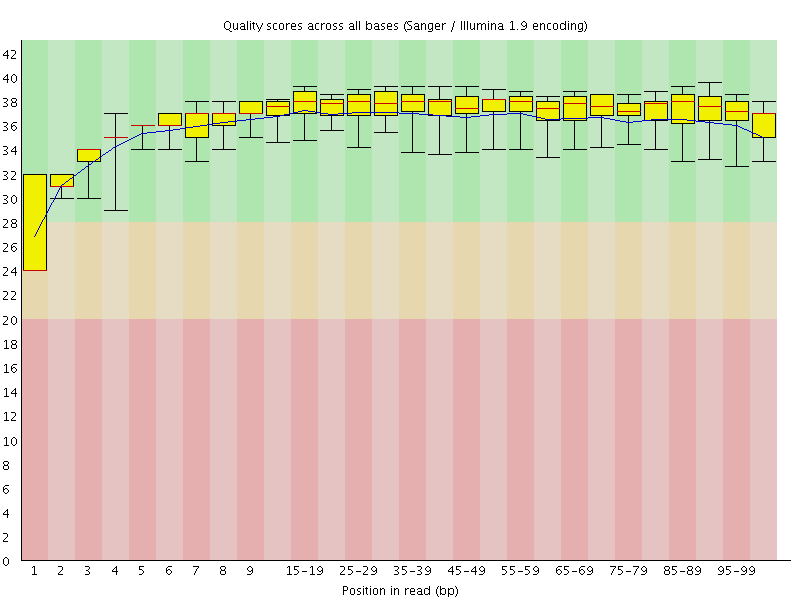
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
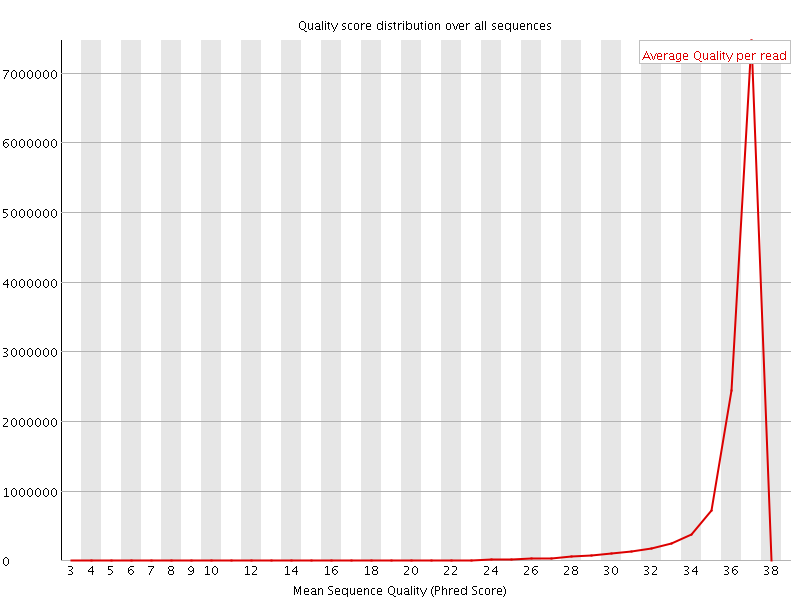
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
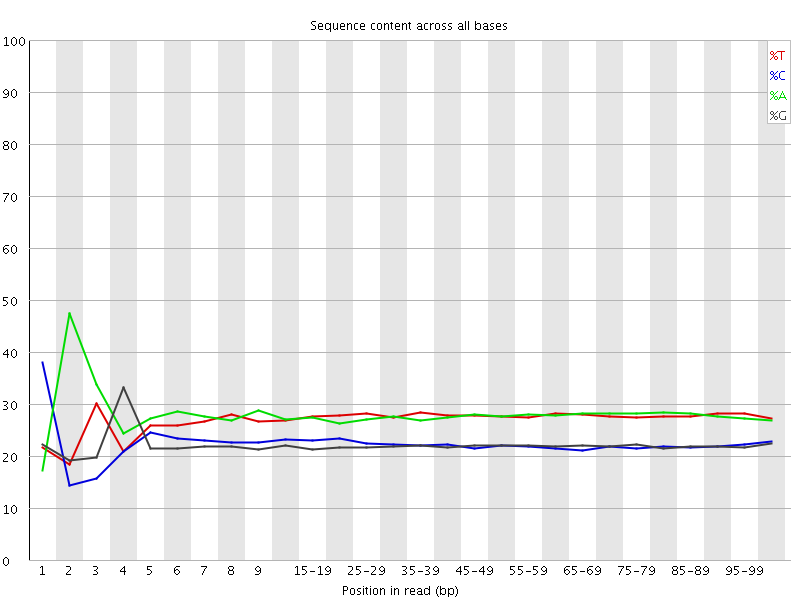
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
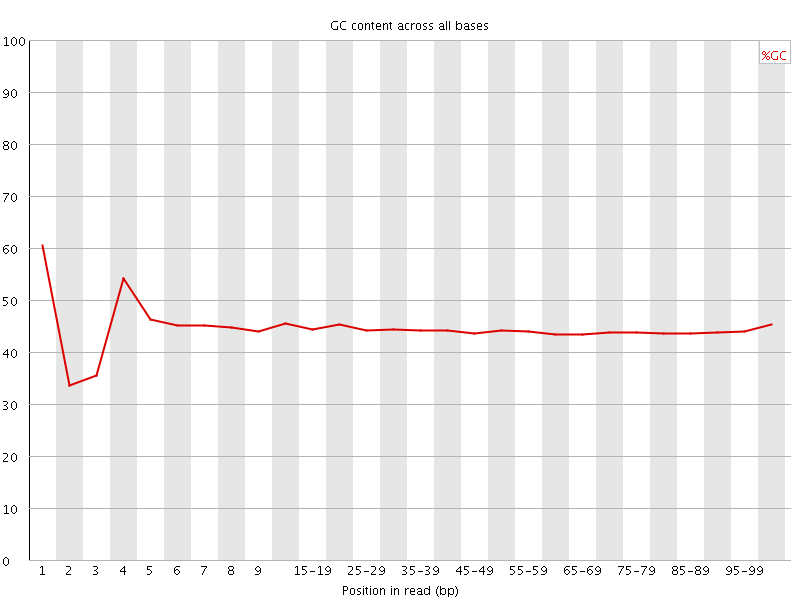
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
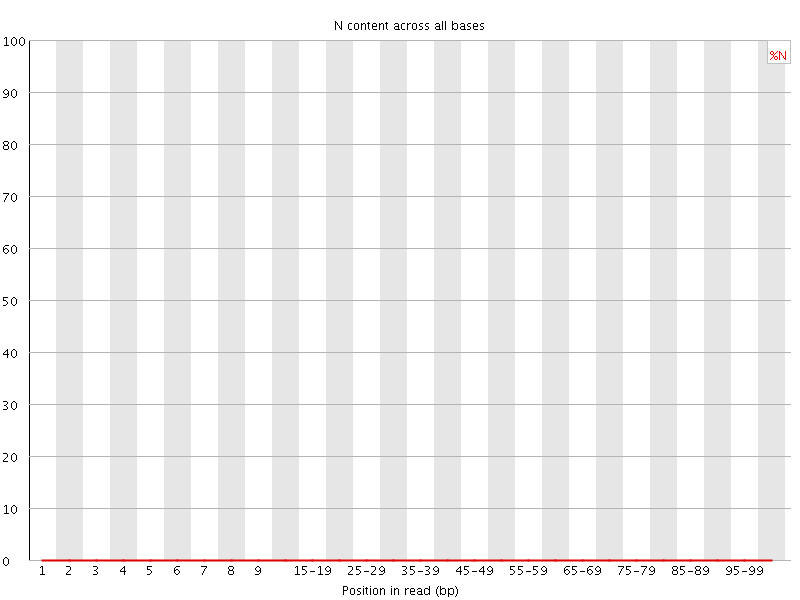
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
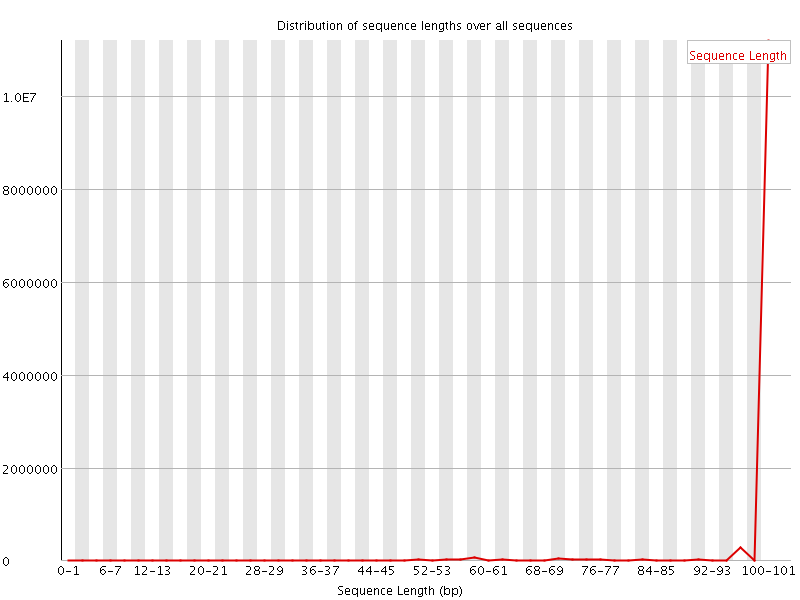
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG | 13443 | 0.11185982033197645 | No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG | 13387 | 0.11139384176033391 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
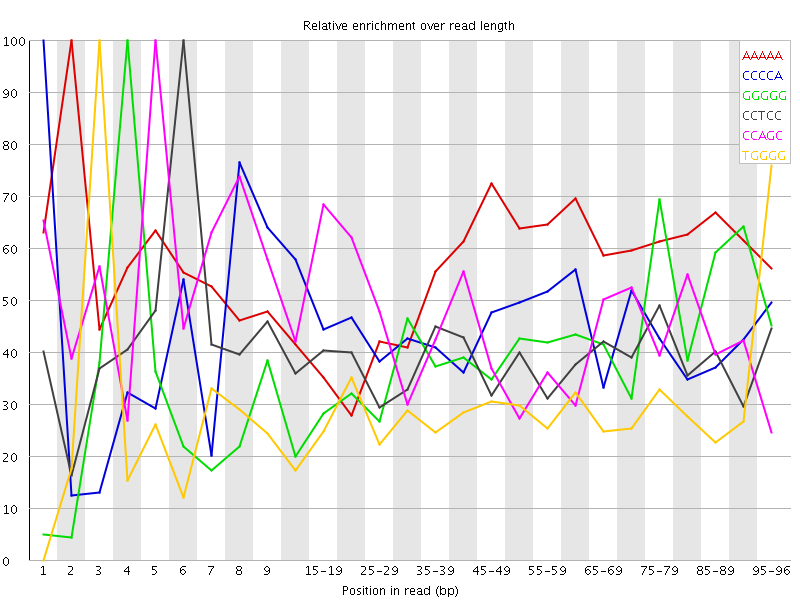
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 5587200 | 2.8943539 | 5.1726093 | 2 |
| CCCCA | 1977340 | 2.5172427 | 5.6406527 | 1 |
| GGGGG | 1490640 | 2.4986365 | 6.221207 | 4 |
| CCTCC | 1895735 | 2.434464 | 6.283842 | 6 |
| CCAGC | 1859145 | 2.390727 | 5.2388415 | 5 |
| TGGGG | 1756850 | 2.3488343 | 8.316749 | 3 |
| CCACC | 1662955 | 2.1170168 | 5.075805 | 1 |
| TGGCT | 1812325 | 1.9132344 | 5.2348447 | 3 |
| TGCAG | 1796395 | 1.8799746 | 6.3977566 | 3 |
| GCCCA | 1412270 | 1.8160781 | 5.613486 | 2 |
| GGCTC | 1346050 | 1.7637334 | 6.8581696 | 1 |
| GCTCC | 1226070 | 1.590428 | 6.212158 | 2 |
| TGCCC | 1218700 | 1.5808678 | 8.33116 | 3 |
| GGCCC | 947285 | 1.5406076 | 5.9813175 | 1 |
| CATGG | 1450830 | 1.5183316 | 36.795895 | 1 |
| TAAAA | 2866850 | 1.4981122 | 7.0831246 | 1 |
| ATGGA | 1766600 | 1.476613 | 10.103628 | 2 |
| TGGCC | 1088305 | 1.4260093 | 5.391006 | 3 |
| TGGGT | 1230920 | 1.3126075 | 5.6024284 | 3 |
| ATGTG | 1550095 | 1.3069795 | 8.894675 | 2 |
| ATGGC | 1222525 | 1.2794043 | 9.343188 | 2 |
| ATGGG | 1185450 | 1.2531594 | 11.318693 | 2 |
| ATGAA | 1878040 | 1.2411915 | 7.7100706 | 2 |
| TGAGA | 1462190 | 1.2221721 | 5.3443384 | 1 |
| ATGCC | 1109425 | 1.1494102 | 7.235652 | 2 |
| CATGT | 1318120 | 1.1002527 | 27.584404 | 1 |
| CATGA | 1315655 | 1.0886734 | 23.646336 | 1 |
| ATGCT | 1274750 | 1.0640512 | 7.7841015 | 2 |
| ATGTT | 1578030 | 1.0612388 | 6.577054 | 2 |
| ATGGT | 1234655 | 1.0410129 | 6.3737864 | 2 |
| ATGAC | 1172430 | 0.9701579 | 6.186356 | 2 |
| CATGC | 932595 | 0.9662071 | 27.77939 | 1 |
| ATGTA | 1374140 | 0.9161084 | 6.327419 | 2 |
| ATGCA | 1047445 | 0.8667358 | 9.384981 | 2 |
| TAAGT | 1109690 | 0.7398056 | 6.3165793 | 1 |
| TAAGG | 824040 | 0.6887741 | 5.192908 | 1 |