| Sequence |
Count |
Percentage |
Possible Source |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
15231 |
0.19013936812459742 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
14637 |
0.18272404512111695 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
13287 |
0.165871038295025 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
13121 |
0.16379874264085367 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
12946 |
0.16161409360784176 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
12887 |
0.16087755479099775 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
11976 |
0.14950489611057569 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
11873 |
0.1482190741082887 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
10759 |
0.13431222254957279 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
10529 |
0.13144096953475715 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
10462 |
0.13060456104783258 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
10420 |
0.13008024527990972 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
10399 |
0.12981808739594827 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
10382 |
0.12960586434702714 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
10174 |
0.12700925292493298 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
10164 |
0.1268844158373323 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
10094 |
0.12601055622412752 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
10069 |
0.1256984635051258 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
9531 |
0.11898222819220916 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
9516 |
0.11879497256080815 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
9484 |
0.11839549388048597 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
9475 |
0.11828314050164536 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
9115 |
0.11378900534802082 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
9076 |
0.11330214070637817 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
9014 |
0.11252815076325397 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
8991 |
0.11224102546177239 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
8931 |
0.11149200293616829 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
8911 |
0.11124232876096693 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
8816 |
0.11005637642876047 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
8808 |
0.10995650675867993 |
No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG |
8707 |
0.10869565217391304 |
No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG |
8678 |
0.10833362461987107 |
No Hit |
| ACAGTGAAAACAGTGACAATGTTCCAGCAAAGCCACCAGACAAAGTTCTT |
8662 |
0.10813388527970996 |
No Hit |
| CATGGCTCTGCTGTGCCTTCCATCCTGGGCTCCCTTCTCTCCTGTGACCT |
8660 |
0.10810891786218985 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
8577 |
0.1070727700351042 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
8558 |
0.1068355795686629 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
8520 |
0.10636119863578031 |
No Hit |
| AGTTGACAACTTGGACTTGGCCAATACTGCATTCTAAATCACCACATCAT |
8467 |
0.10569956207149668 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
8458 |
0.10558720869265609 |
No Hit |
| TAATCTGATGTGGCATTTTCGTCATCTGAAGCATGAGTGACAAGTTGGGA |
8436 |
0.1053125670999346 |
No Hit |
| CATGGGGTGGGTGCTTGCAAGTGTAAGCCTCGGCAAACAAGGCCTGGCTT |
8168 |
0.10196693315223633 |
No Hit |
| GGCTGCTTGTTGTACGTGGCCTGGTGGAACGCACTGCAAAACGAGCTCAG |
8048 |
0.10046888810102815 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality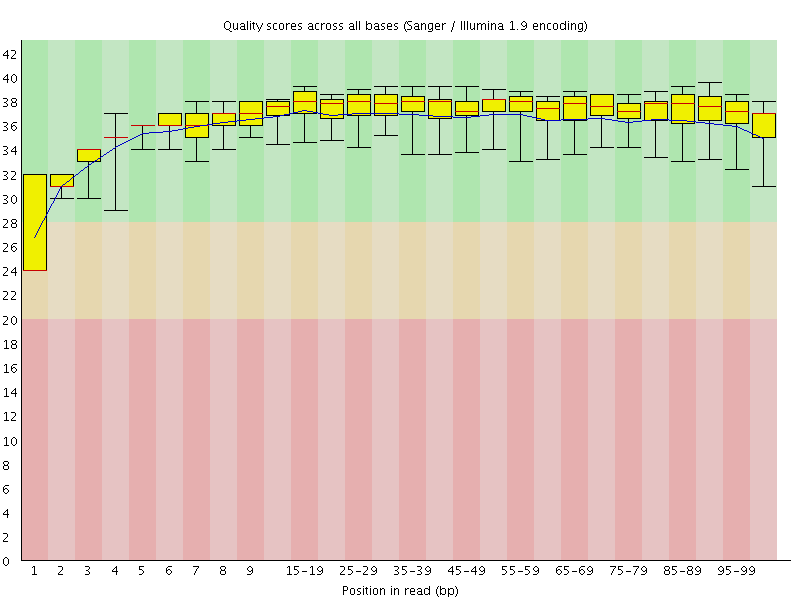
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores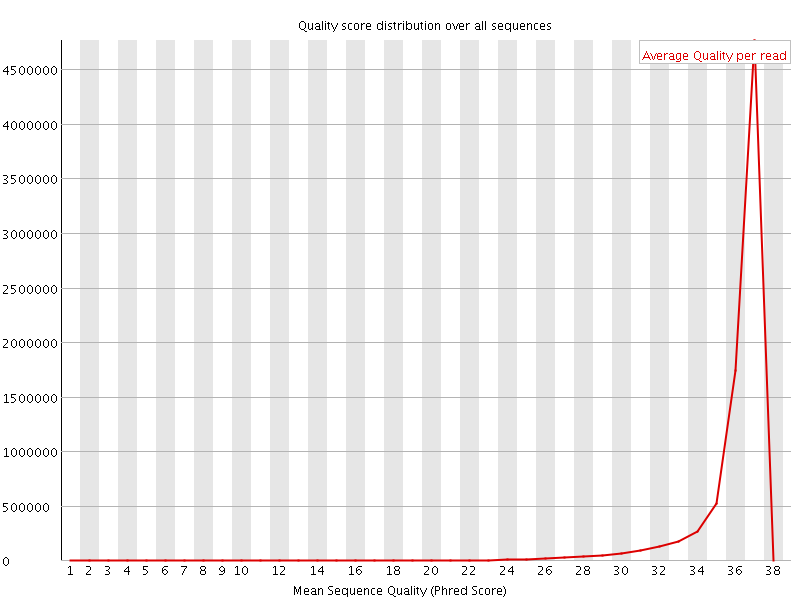
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content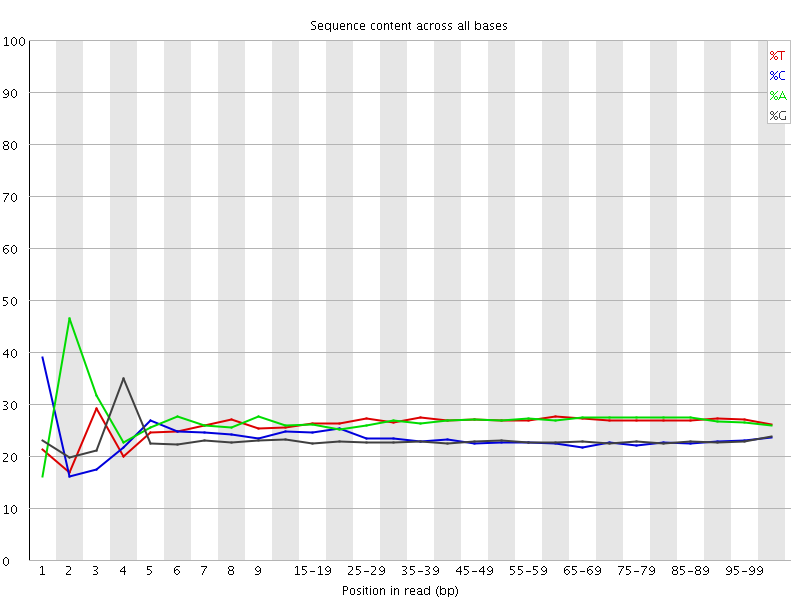
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content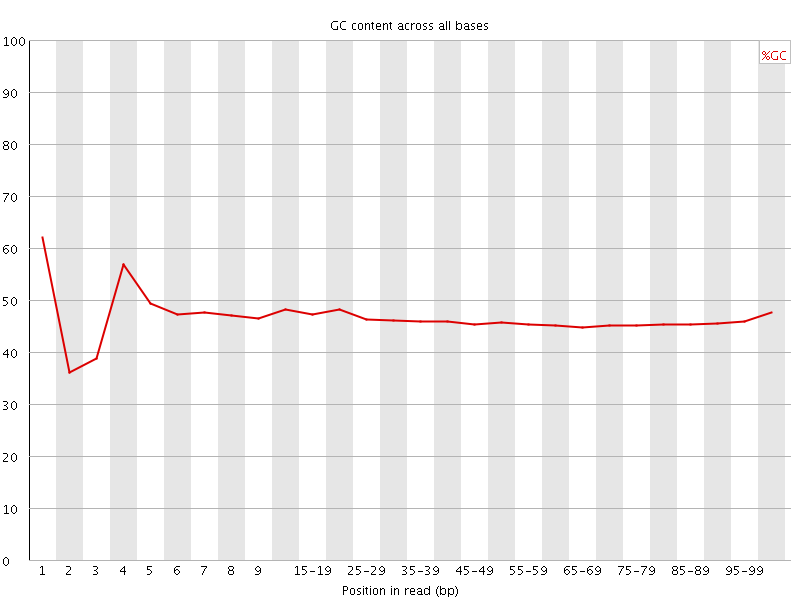
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content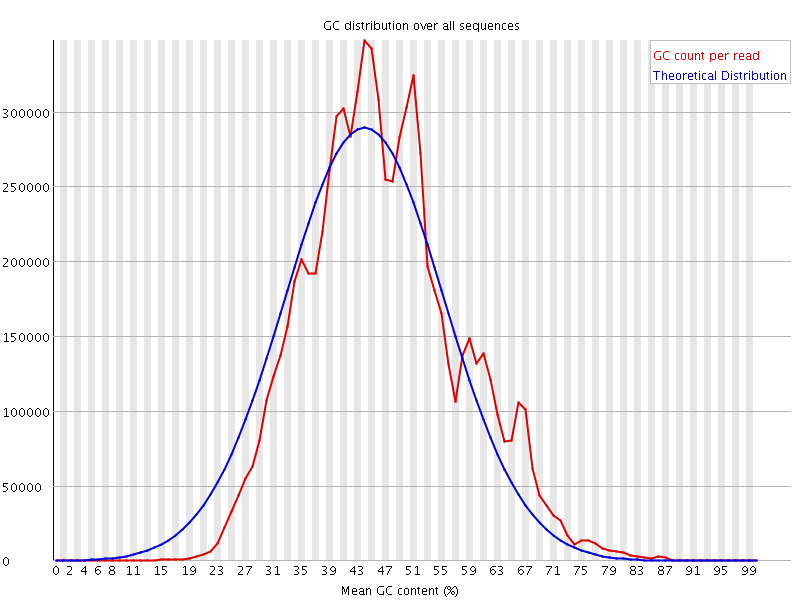
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution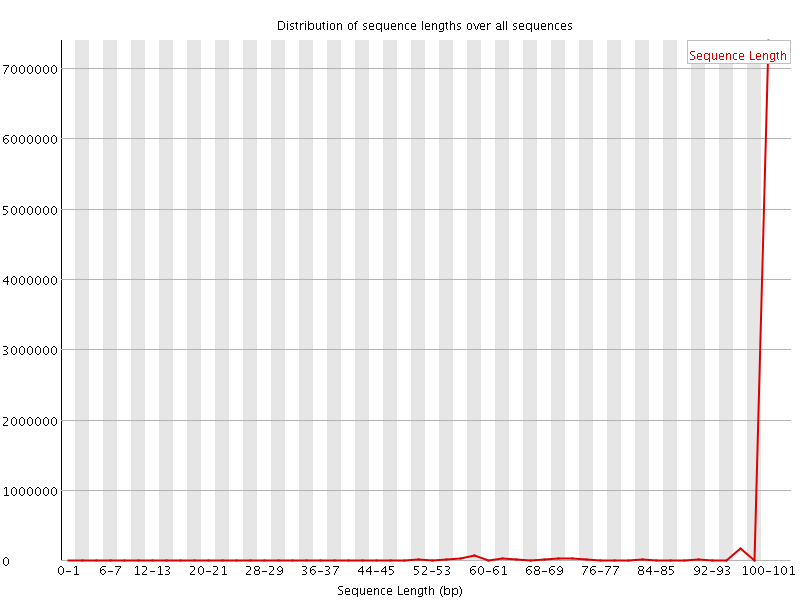
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels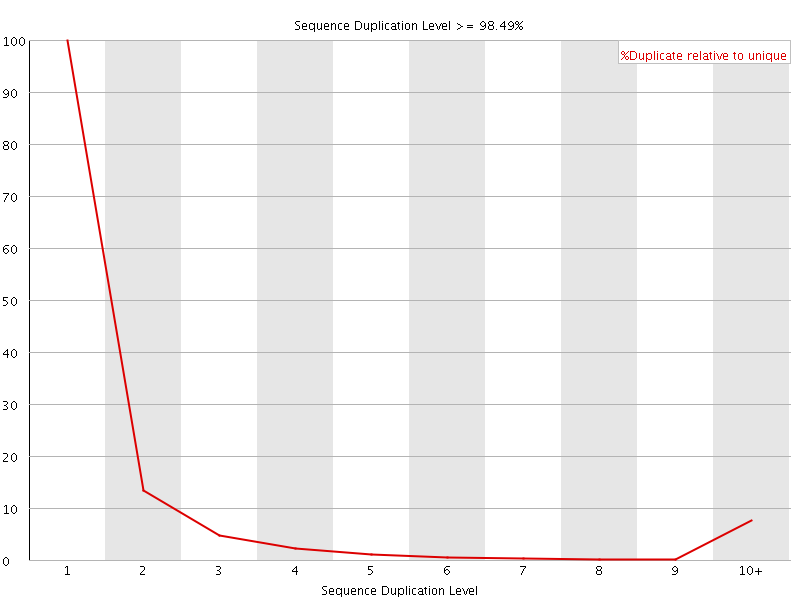
![[WARN]](Icons/warning.png) Overrepresented sequences
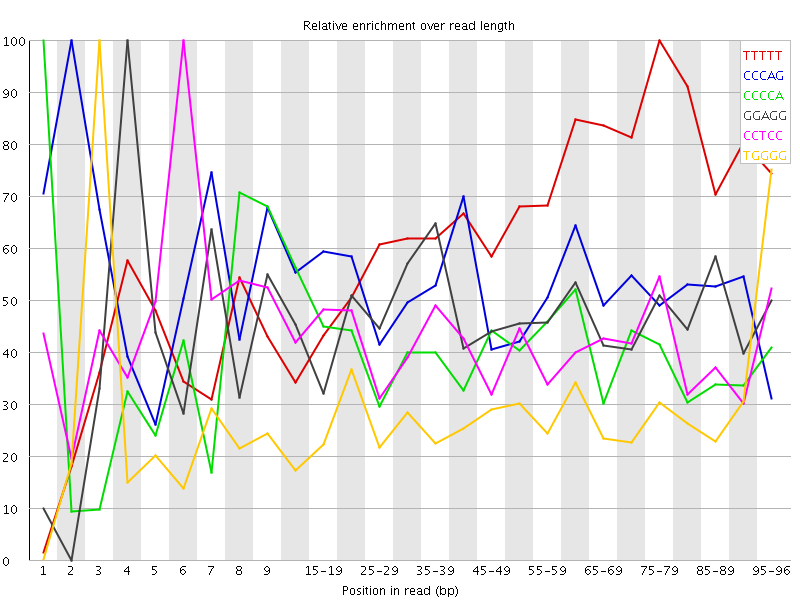
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content