![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-37.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 8598466 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 45 |
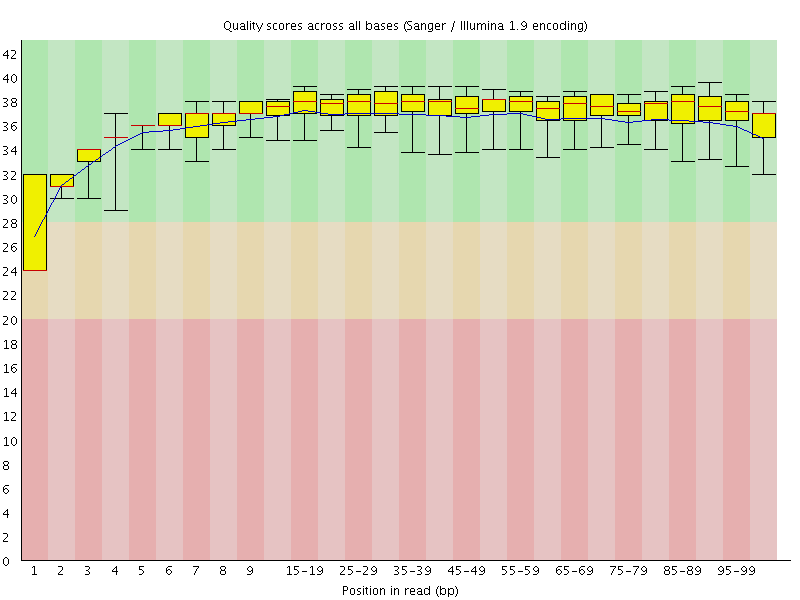
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
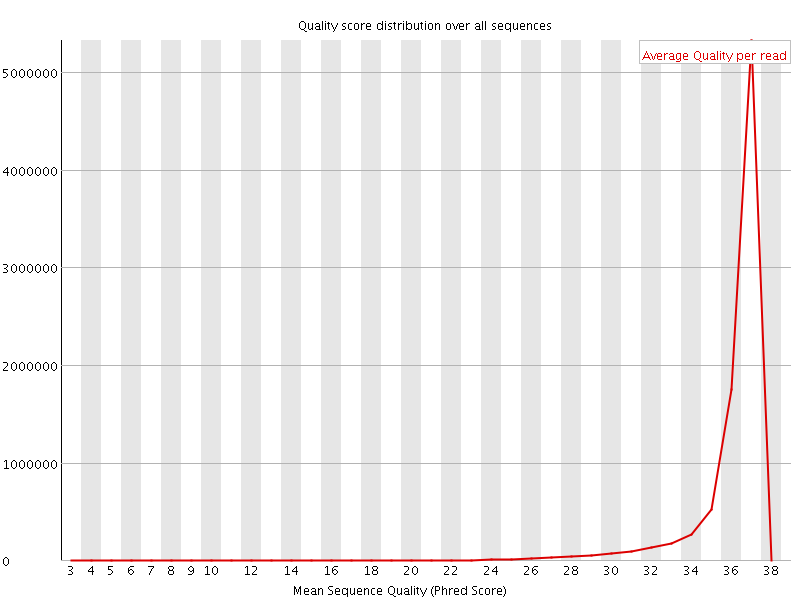
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
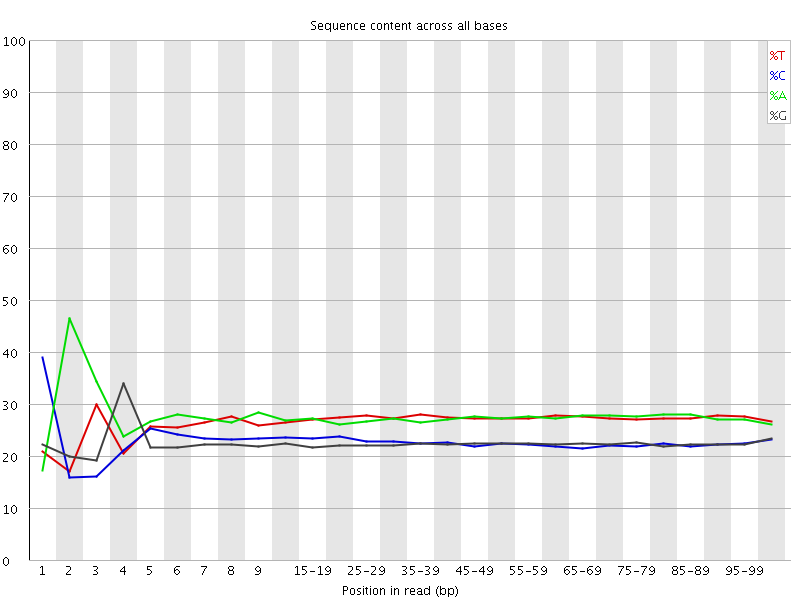
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
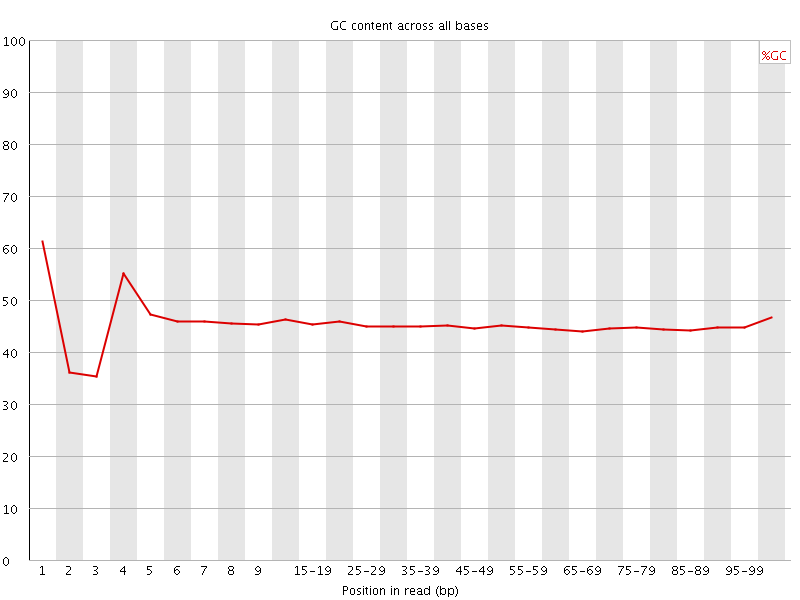
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
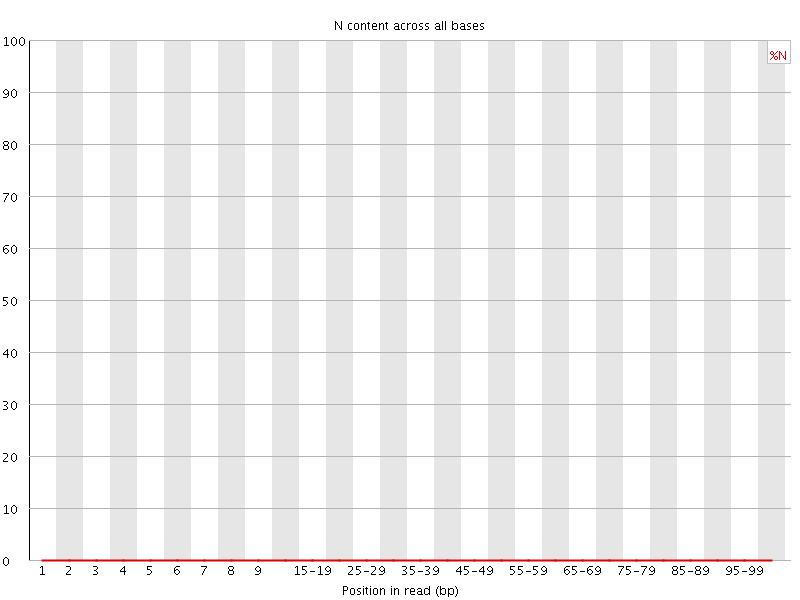
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
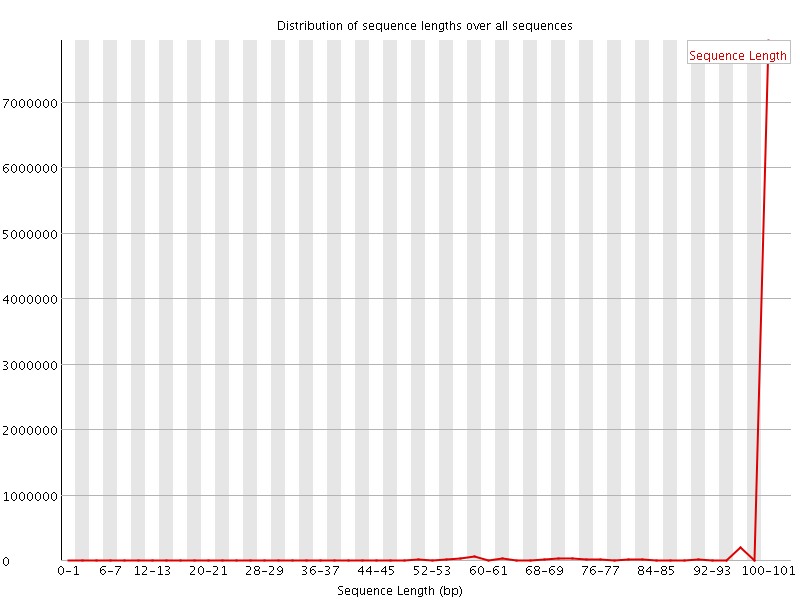
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 11464 | 0.13332610723819807 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 11098 | 0.1290695340308376 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 10866 | 0.12637137833655446 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 10855 | 0.12624344854070482 | No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG | 10818 | 0.12581313922739243 | No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG | 10773 | 0.125289790062553 | No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA | 9995 | 0.1162416645015518 | No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT | 9940 | 0.11560201552230362 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 9914 | 0.11529963600484087 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 9882 | 0.11492747659873286 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 9816 | 0.11415989782363506 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 9778 | 0.11371795852888178 | No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA | 9412 | 0.1094613853215213 | No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA | 9375 | 0.1090310760082089 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 8664 | 0.10076215920374633 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 8650 | 0.10059933946357409 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
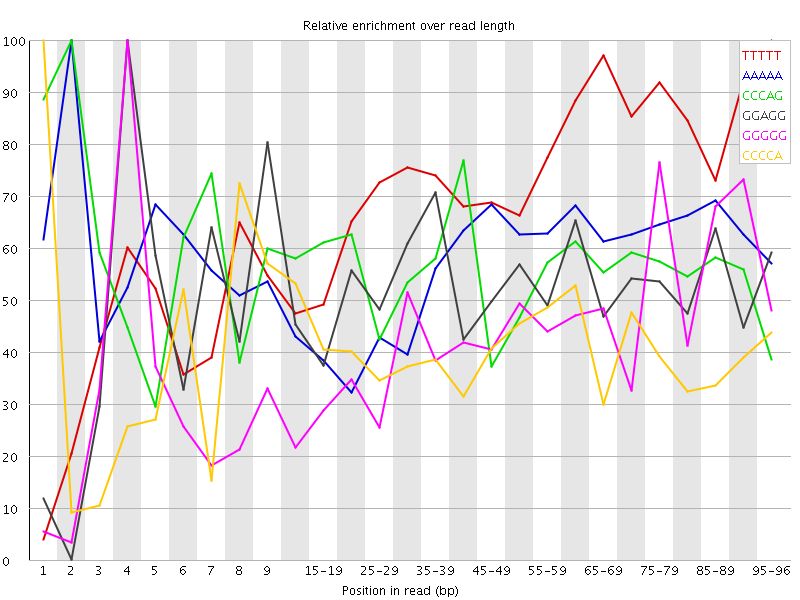
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 3838560 | 3.1404827 | 4.340371 | 95-96 |
| AAAAA | 3954825 | 3.0750792 | 5.383879 | 2 |
| CCCAG | 1708675 | 2.907213 | 5.1420274 | 2 |
| GGAGG | 1497740 | 2.6353402 | 5.049282 | 4 |
| GGGGG | 1189870 | 2.5678375 | 5.8929605 | 4 |
| CCCCA | 1515240 | 2.5493994 | 6.2802076 | 1 |
| CCTCC | 1441600 | 2.45031 | 5.732795 | 6 |
| TGGGG | 1342910 | 2.3870802 | 8.6180725 | 3 |
| CCACC | 1267665 | 2.132853 | 5.7519655 | 1 |
| TGGCT | 1320445 | 1.9117515 | 6.180281 | 3 |
| TGCAG | 1284265 | 1.8405429 | 6.387155 | 3 |
| GCCCA | 1073545 | 1.8265754 | 5.808334 | 2 |
| GGCTC | 1038045 | 1.8043234 | 6.579027 | 1 |
| GCTCC | 947135 | 1.62798 | 6.224154 | 2 |
| TGCCC | 927905 | 1.5949267 | 8.820272 | 3 |
| TAAAA | 2017105 | 1.5844455 | 7.112305 | 1 |
| GGCCC | 728300 | 1.5198319 | 6.0382924 | 1 |
| CATGG | 1041575 | 1.492732 | 36.641476 | 1 |
| ATGGA | 1217930 | 1.4391525 | 10.034484 | 2 |
| TCAGG | 998995 | 1.4317086 | 5.358905 | 1 |
| TGGGT | 895535 | 1.3111569 | 5.5495915 | 3 |
| ATGTG | 1094020 | 1.305959 | 9.508759 | 2 |
| ATGGG | 881535 | 1.277591 | 11.612711 | 2 |
| ATGGC | 881185 | 1.2628692 | 10.206316 | 2 |
| ATGAA | 1244490 | 1.1989692 | 7.5789113 | 2 |
| TGAGA | 1012760 | 1.1967158 | 5.684038 | 1 |
| ATGCC | 802100 | 1.1367341 | 6.822869 | 2 |
| TGAGC | 788875 | 1.1305753 | 5.082582 | 1 |
| CATGT | 917950 | 1.0835835 | 28.659557 | 1 |
| ATGTT | 1095070 | 1.0767086 | 7.0323095 | 2 |
| ATGCT | 877520 | 1.0358583 | 8.192999 | 2 |
| CATGA | 885340 | 1.0345073 | 22.711136 | 1 |
| ATGGT | 833960 | 0.99551886 | 6.382676 | 2 |
| ATGAC | 820400 | 0.95862585 | 6.0940943 | 2 |
| CATGC | 656840 | 0.93087196 | 27.800396 | 1 |
| ATGTA | 937595 | 0.91253966 | 6.860658 | 2 |
| ATGCA | 729480 | 0.8523871 | 9.905174 | 2 |
| TAAGT | 775475 | 0.7547519 | 7.1189876 | 1 |
| TAAGG | 573905 | 0.6781479 | 5.04177 | 1 |