| Sequence |
Count |
Percentage |
Possible Source |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
15540 |
0.17714452599476227 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
14984 |
0.1708065365190166 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
14416 |
0.1643317559035066 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
14343 |
0.16349960980327383 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
13560 |
0.15457398793365357 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
13386 |
0.15259051640707127 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11963 |
0.1363693670833553 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11959 |
0.13632377003676718 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
11937 |
0.13607298628053263 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
11746 |
0.13389572730595095 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
11622 |
0.1324822188617199 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
11602 |
0.1322542336287794 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
11561 |
0.13178686390125138 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
11542 |
0.13157027792995793 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
10852 |
0.12370478739351094 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
10806 |
0.12318042135774782 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
10649 |
0.12139073727916495 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
10643 |
0.1213223417092828 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
10228 |
0.1165916481257676 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10183 |
0.11607868135165147 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
10165 |
0.11587349464200504 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10147 |
0.1156683079323586 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
10064 |
0.11472216921565558 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
10053 |
0.1145967773375383 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
9473 |
0.10798520558226402 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
9429 |
0.10748363806979495 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
9369 |
0.10679968237097347 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
9317 |
0.10620692076532819 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
9315 |
0.10618412224203415 |
No Hit |
| ACAGTGAAAACAGTGACAATGTTCCAGCAAAGCCACCAGACAAAGTTCTT |
9285 |
0.1058421443926234 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
9252 |
0.10546596875827159 |
No Hit |
| CATGGCTCTGCTGTGCCTTCCATCCTGGGCTCCCTTCTCTCCTGTGACCT |
9230 |
0.10521518500203705 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
9109 |
0.10383587434274708 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
9099 |
0.10372188172627682 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
8893 |
0.10137363382698977 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
8867 |
0.10107725302416712 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
8834 |
0.1007010773898153 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
8798 |
0.10029070397052242 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality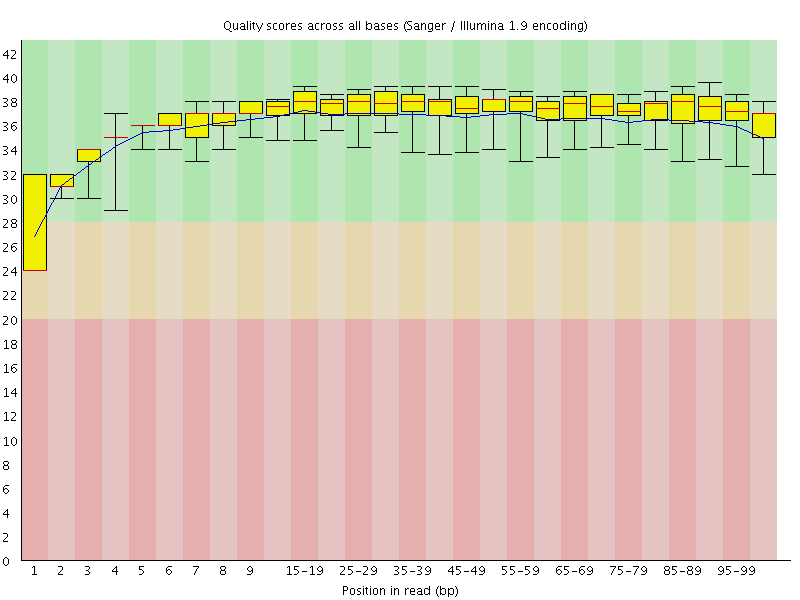
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores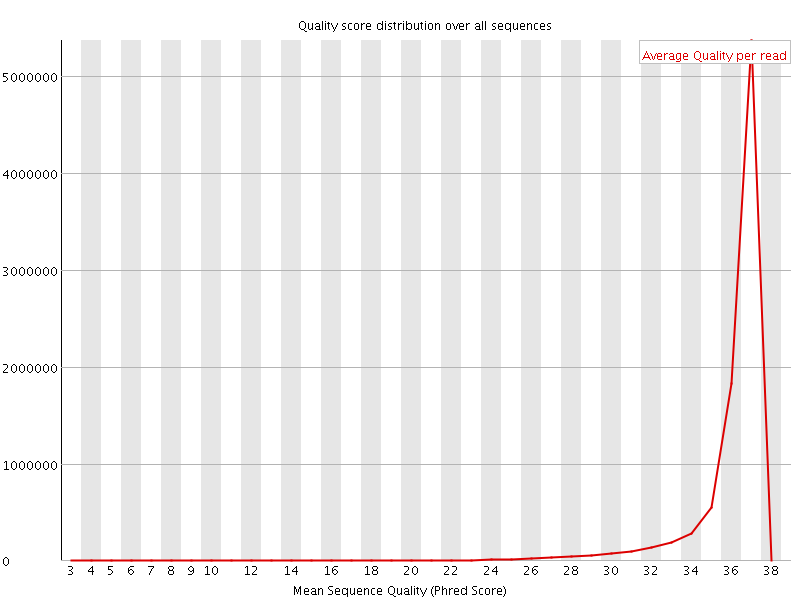
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content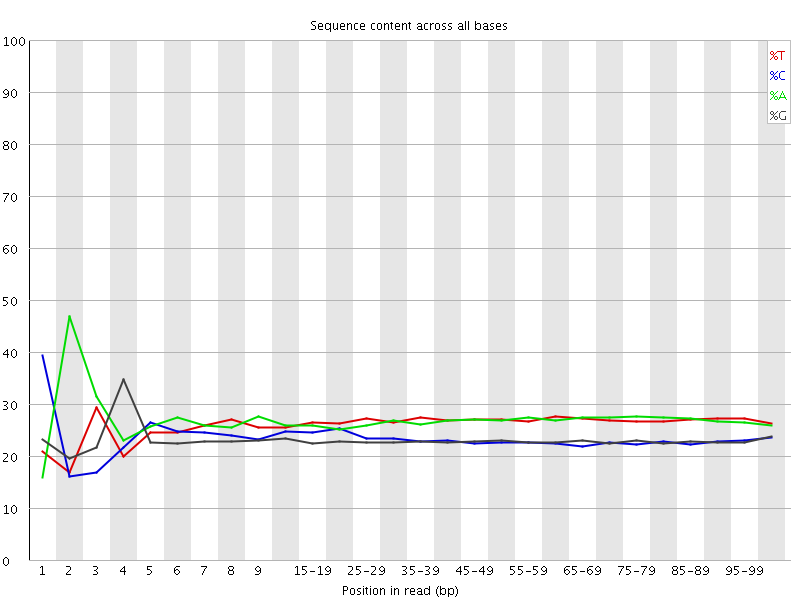
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content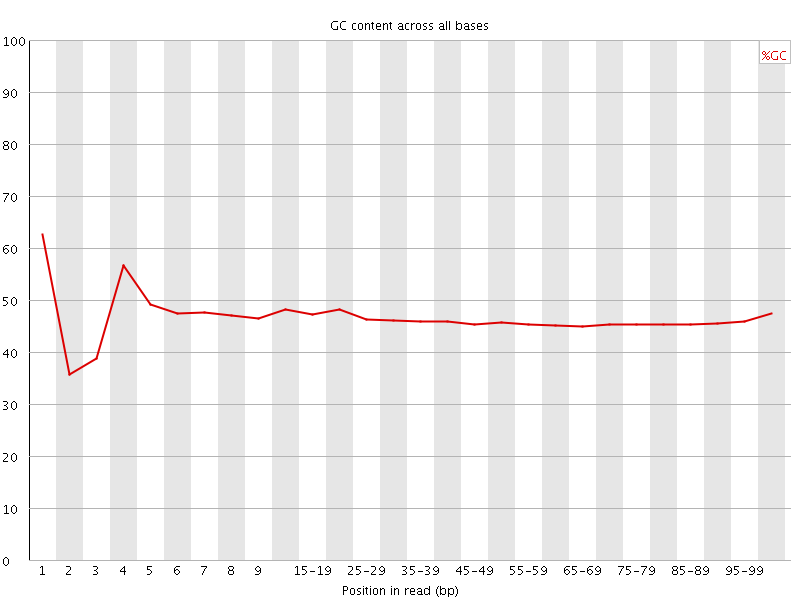
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content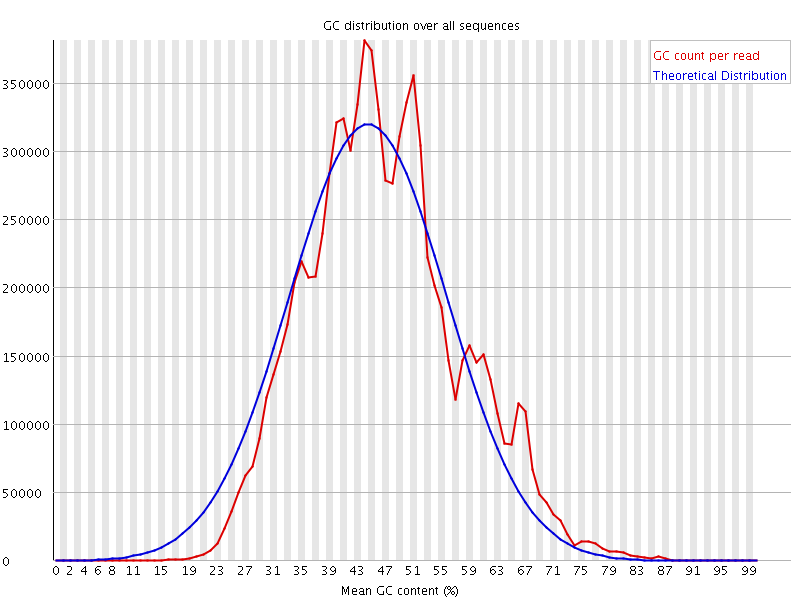
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution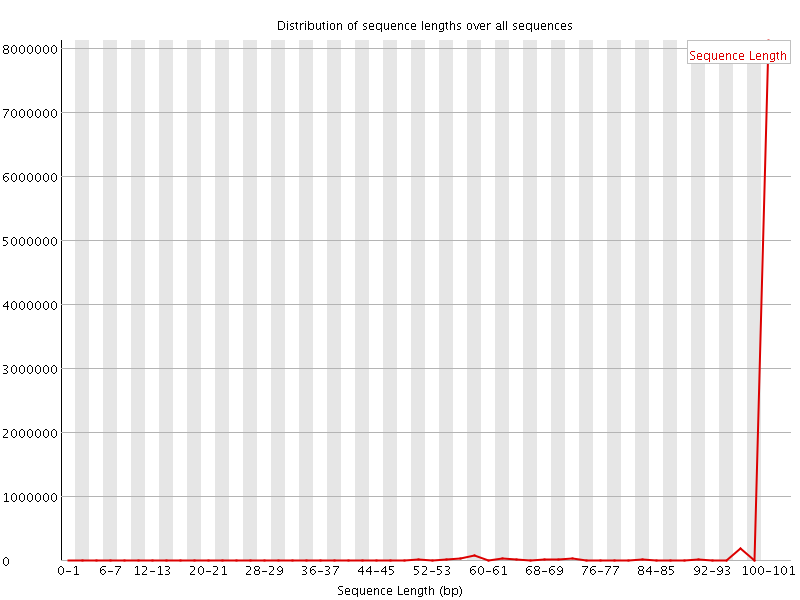
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels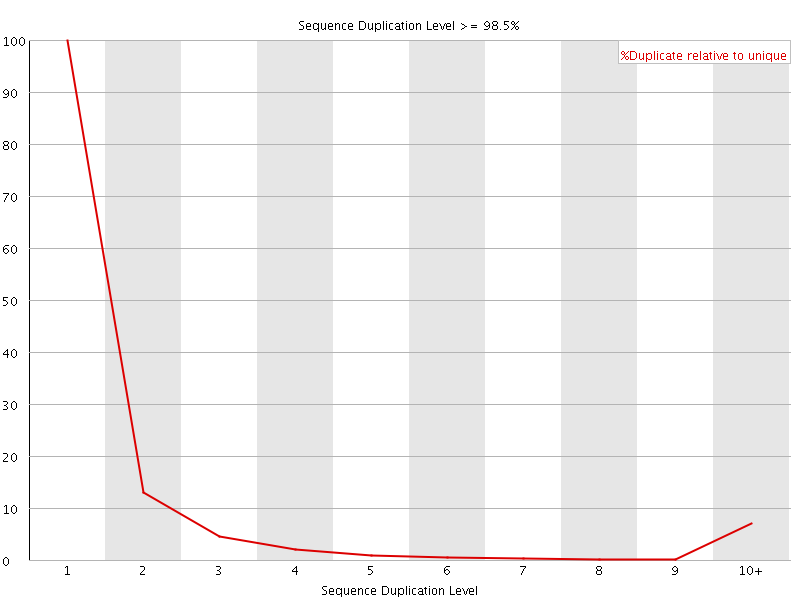
![[WARN]](Icons/warning.png) Overrepresented sequences
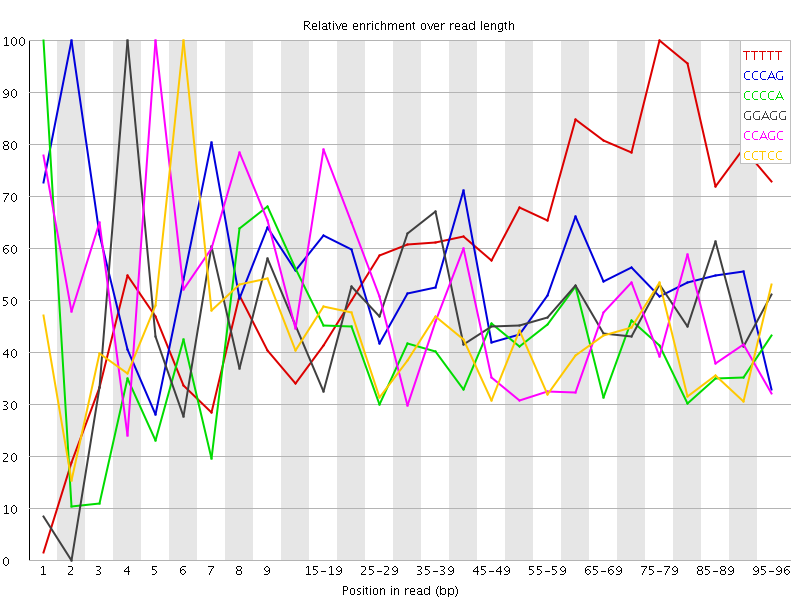
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content