| Sequence |
Count |
Percentage |
Possible Source |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
15139 |
0.17547511809420968 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
15081 |
0.1748028440437794 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
14136 |
0.16384941339452727 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
13649 |
0.15820462955729364 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
11581 |
0.13423458237988262 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
11529 |
0.13363185392087612 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
11393 |
0.13205548718193613 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
11275 |
0.1306877572172676 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
11138 |
0.12909979954642364 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
10984 |
0.1273147960332122 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
10654 |
0.12348978850490193 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
10627 |
0.12317683334349472 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
10612 |
0.12300296936493516 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
10572 |
0.12253933208877633 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
10236 |
0.11864477896904224 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
10155 |
0.11770591348482062 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10125 |
0.11735818552770151 |
No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG |
10106 |
0.11713795782152608 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10097 |
0.11703363943439034 |
No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG |
10091 |
0.11696409384296652 |
No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC |
9724 |
0.11271022183420934 |
No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA |
9721 |
0.11267544903849741 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
9627 |
0.1115859014395242 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
9614 |
0.11143521932477259 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
9374 |
0.10865339566781965 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
9374 |
0.10865339566781965 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
9245 |
0.10715816545220747 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
9220 |
0.10686839215460821 |
No Hit |
| AGTTGACAACTTGGACTTGGCCAATACTGCATTCTAAATCACCACATCAT |
9163 |
0.10620770903608187 |
No Hit |
| TAATCTGATGTGGCATTTTCGTCATCTGAAGCATGAGTGACAAGTTGGGA |
9069 |
0.10511816143710866 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
8699 |
0.10082951663263956 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
8647 |
0.10022678817363309 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality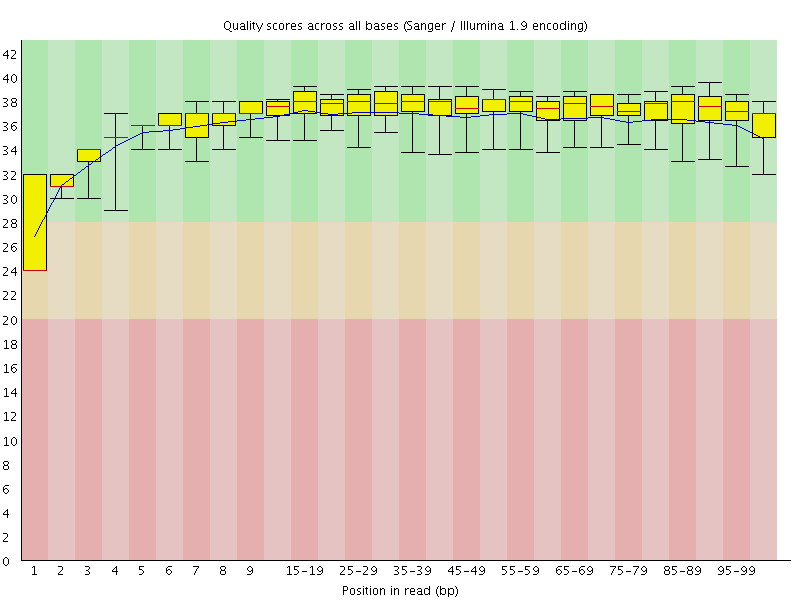
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores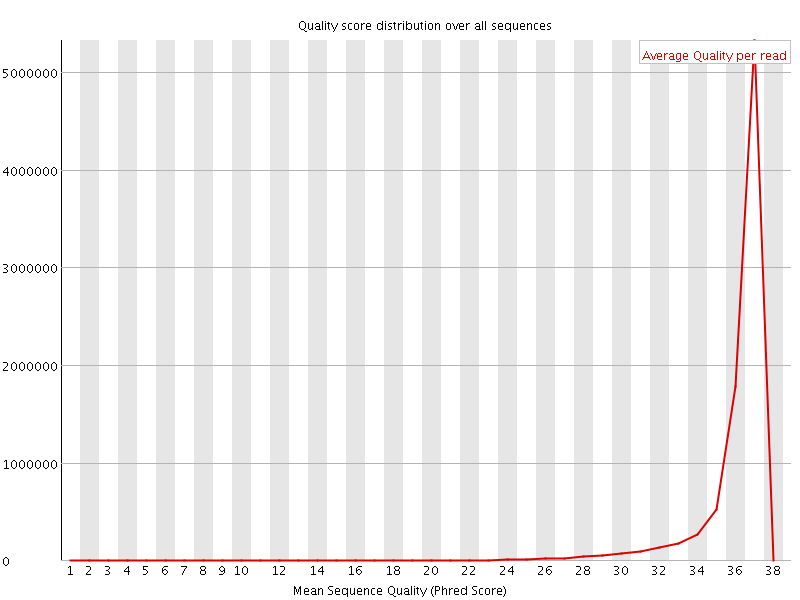
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content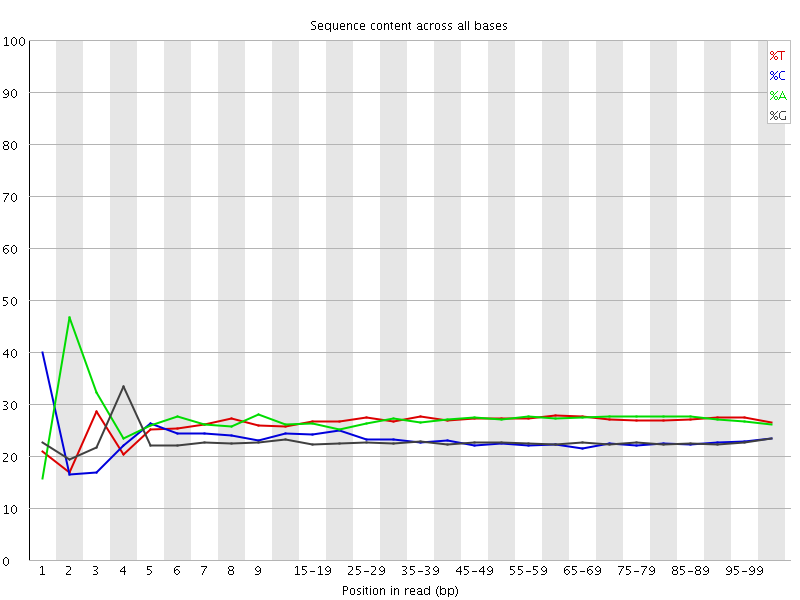
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content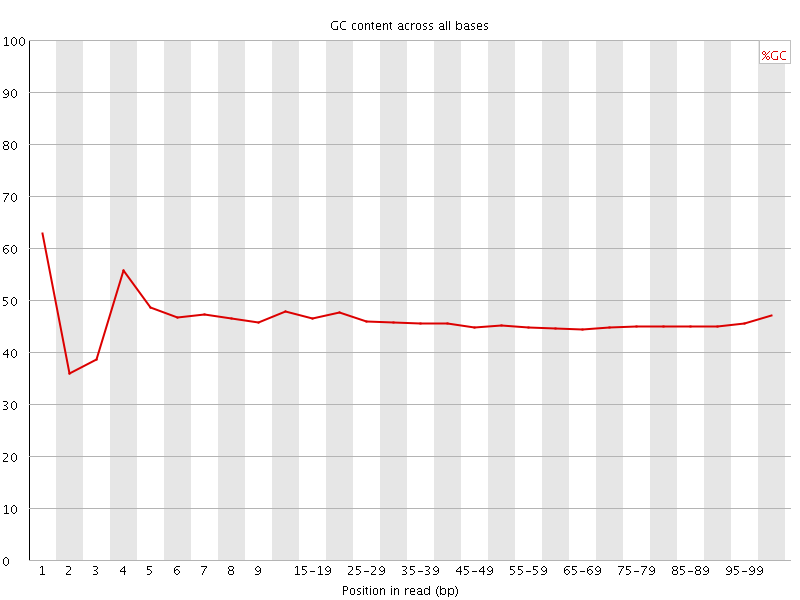
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content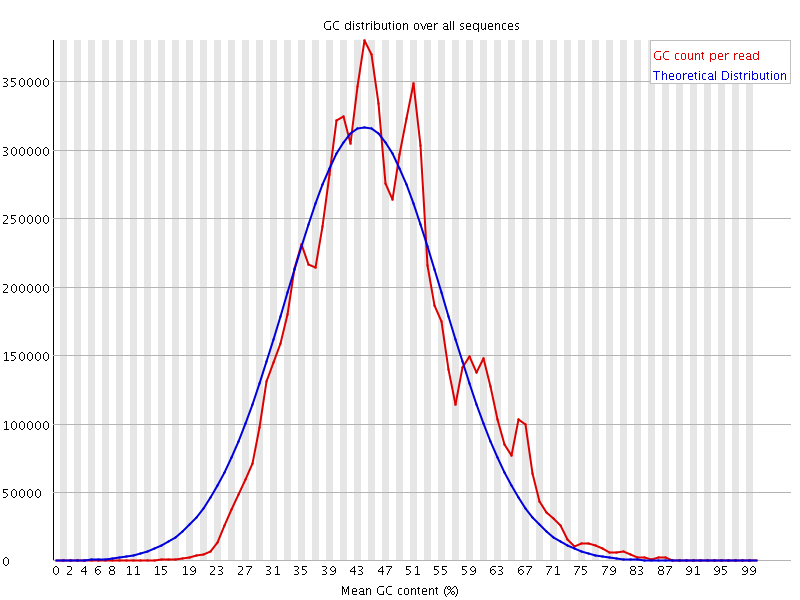
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution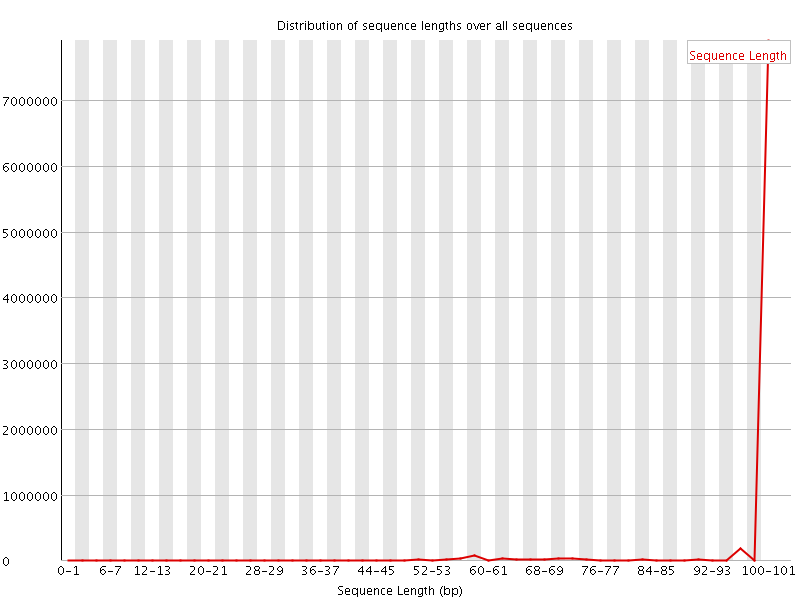
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels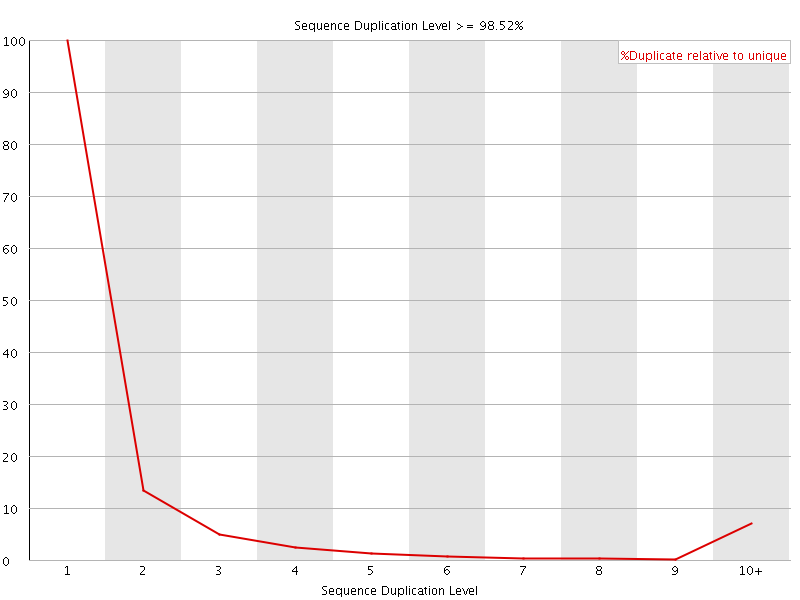
![[WARN]](Icons/warning.png) Overrepresented sequences
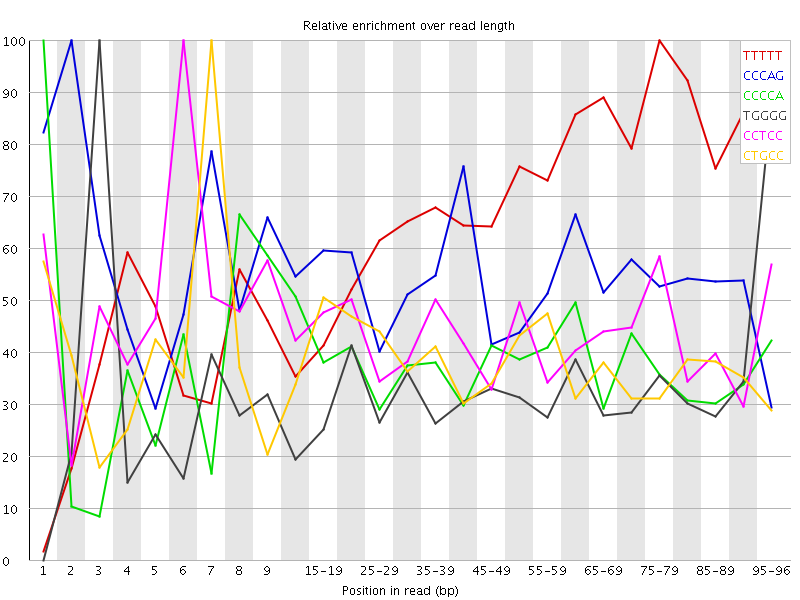
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content