| Sequence |
Count |
Percentage |
Possible Source |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
18429 |
0.2072987051845486 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
18387 |
0.20682626795964484 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
17287 |
0.1944529120693088 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
17150 |
0.19291186683569422 |
No Hit |
| AGAAGAGGGCTCCTTTTGTGCATATCAAACCTTGTTCCAGAATGGAGTGG |
15089 |
0.16972869729934634 |
No Hit |
| GACCCCCTCCACTAACTCCCGAGGACGTTGGCTTTGCATCTGGTTTTTCT |
15031 |
0.16907628398876498 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
14896 |
0.1675577357658601 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
14832 |
0.16683783142314965 |
No Hit |
| GATCCAAAACTTCTCTCCGACACTGAATACCTTCTGCTTCTCCTATCATT |
14642 |
0.16470061540572797 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
14428 |
0.16229343525978984 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
14412 |
0.16211345917411224 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
14313 |
0.160999857143982 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
13931 |
0.15670292809842892 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
13066 |
0.14697297096648285 |
No Hit |
| GACCACACACCTTGGCTTATGCCTCCAACAATCCCCTTAGCATCACCATT |
13065 |
0.14696172246112799 |
No Hit |
| TGACCTAGAATGCTTTAAAGAAGTCCACCTAAATGTCGGTTCTCGCAAAA |
13011 |
0.14635430317196602 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
12856 |
0.14461078484196413 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11628 |
0.13079762026620714 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11622 |
0.130730129234078 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
11392 |
0.1281429730024623 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
11326 |
0.12740057164904214 |
No Hit |
| CATGAACTGCCTGTCAACAGCTAACTATTTATGTAAATCAGTCTTACCAG |
10850 |
0.12204628310013307 |
No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA |
10755 |
0.12097767509142224 |
No Hit |
| CAGAAGCTGCACTTTATAAGAATCTTCTGCATTCAAAGGAGTCTTCCTGG |
10742 |
0.12083144452180916 |
No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC |
10576 |
0.1189641926329039 |
No Hit |
| CATGCTTTCCCTGGGTCTTCACGGATTGCTTTCCAAGCTGCCTTGTTGCG |
10482 |
0.1179068331295479 |
No Hit |
| GCGACAGCTGGAAAGGTCCAGTTGCTGCTCTGCAGCAACCCCGCAACAAG |
10410 |
0.11709694074399865 |
No Hit |
| CATGGAAAGCCCCTGTTTCATACTGACCAAAACTCAGCCTGTTTCTGGGA |
10349 |
0.11641078191735274 |
No Hit |
| TATGAAGTACAGTGGAAGGTTGTTGAGGAGATAAATGGAAACAATTATGT |
10306 |
0.11592709618709414 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
10042 |
0.11295749077341348 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
10028 |
0.11280001169844556 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
10027 |
0.11278876319309072 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
9973 |
0.11218134390392877 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
9874 |
0.11106774187379853 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
9840 |
0.11068529269173358 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
9831 |
0.11058405614353993 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
9821 |
0.11047157108999142 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
9794 |
0.11016786144541044 |
No Hit |
| TGATTTTTCTCTGCAGATTCCCCAGACTGAGTCAGGGAATGAGAAGGAAA |
9744 |
0.10960543617766788 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
9737 |
0.10952669664018393 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
9584 |
0.10780567532089173 |
No Hit |
| GAGCTCTCACCTCTCACCCCCCCGTCTTCTGTCTCTTCCTCGTTAAGCAT |
9471 |
0.10653459421579357 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
9457 |
0.10637711514082566 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
9415 |
0.10590467791592192 |
No Hit |
| GAAGGGCAGGAGTTCCTTTGGTGACTTCACAGTGAAGTCTTGCCCTCTCT |
9274 |
0.10431863866088795 |
No Hit |
| TAGCTGCTCCATTTTCATTTCACCACTTTGAAATCTTGGAACCACTGATG |
9272 |
0.10429614165017823 |
No Hit |
| TAAGTATACCTTGGCTTCGTTCAGTTATAATTTCAACATGTATGGTTGTT |
9045 |
0.10174273093462706 |
No Hit |
| ATCCGGGGTCCTCGCACAGAGGTCTCTGATGAGTCACTTTCTTGACCCTG |
9024 |
0.1015065123221752 |
No Hit |
| CATGGGGTGGGTGCTTGCAAGTGTAAGCCTCGGCAAACAAGGCCTGGCTT |
8954 |
0.10071911694733562 |
No Hit |
| GGCTGCTTGTTGTACGTGGCCTGGTGGAACGCACTGCAAAACGAGCTCAG |
8898 |
0.10008920064746396 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality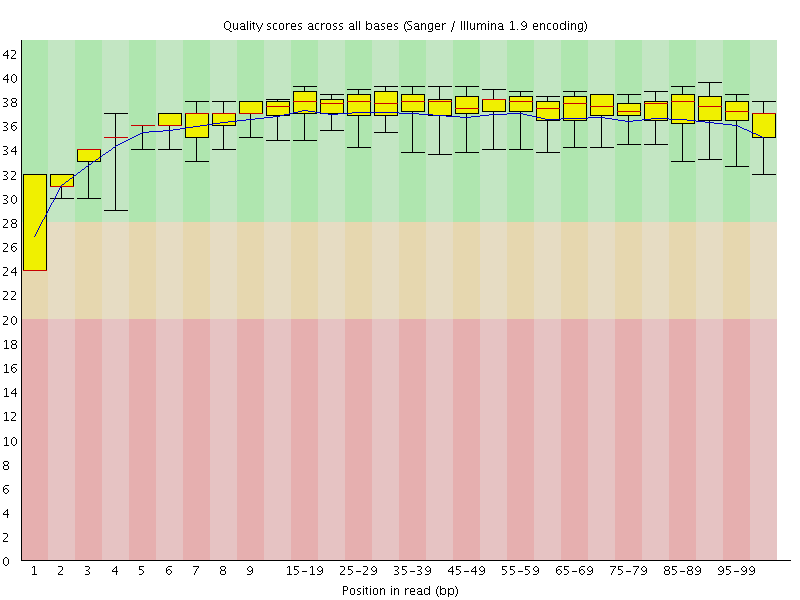
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores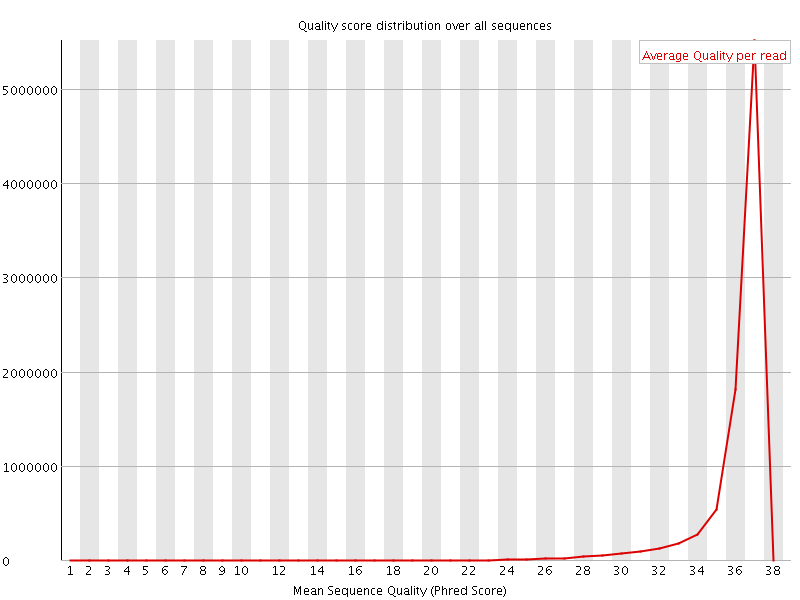
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content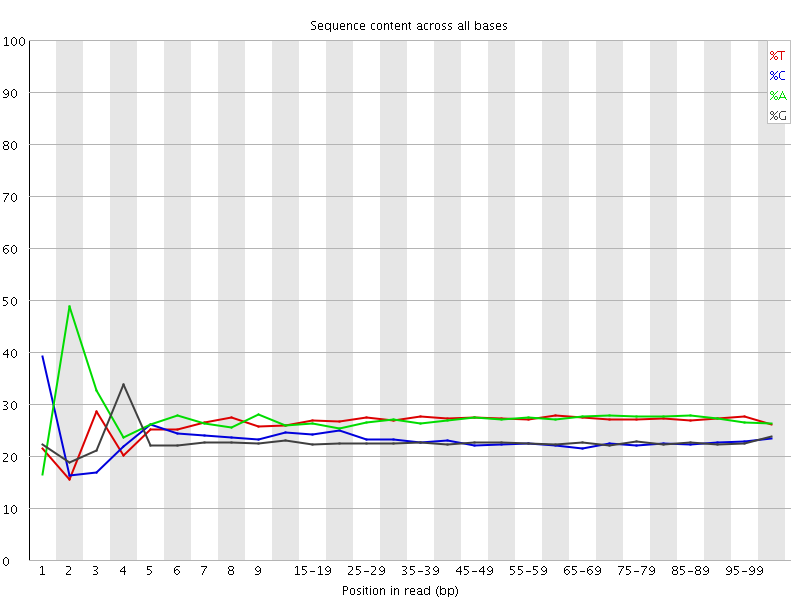
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content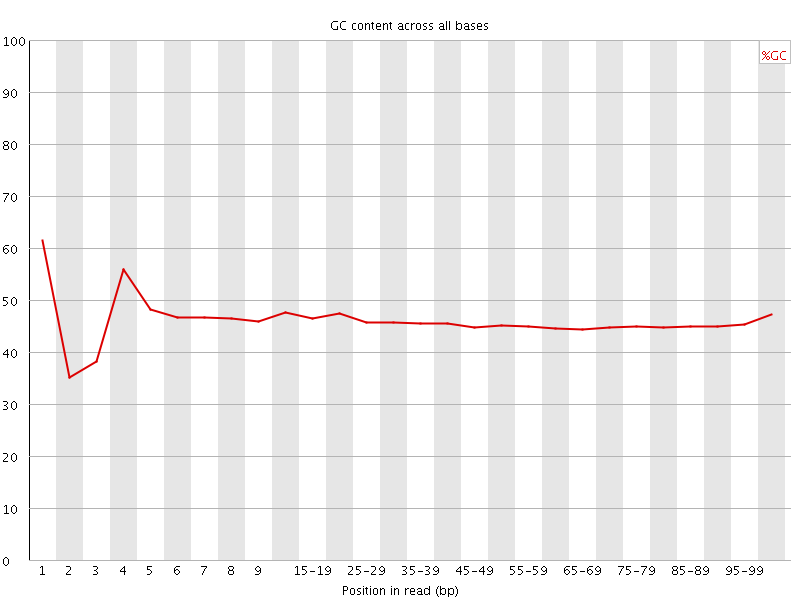
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content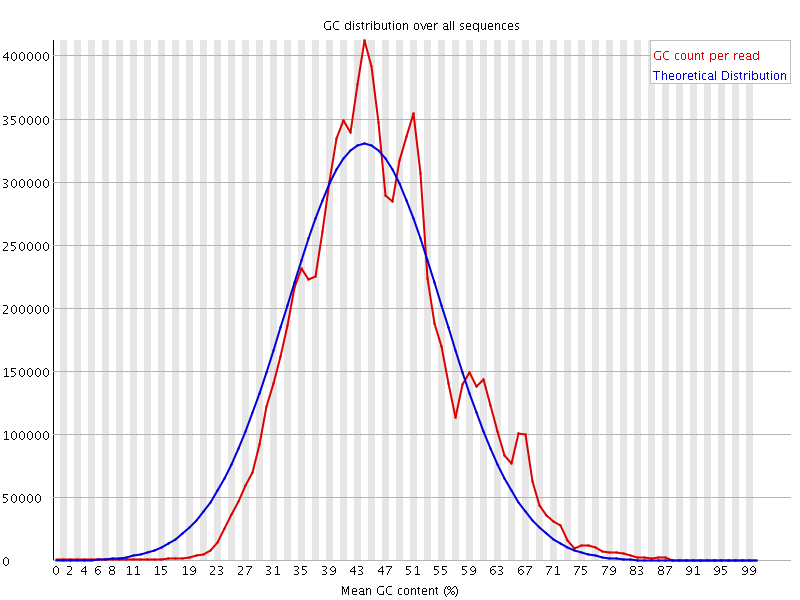
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution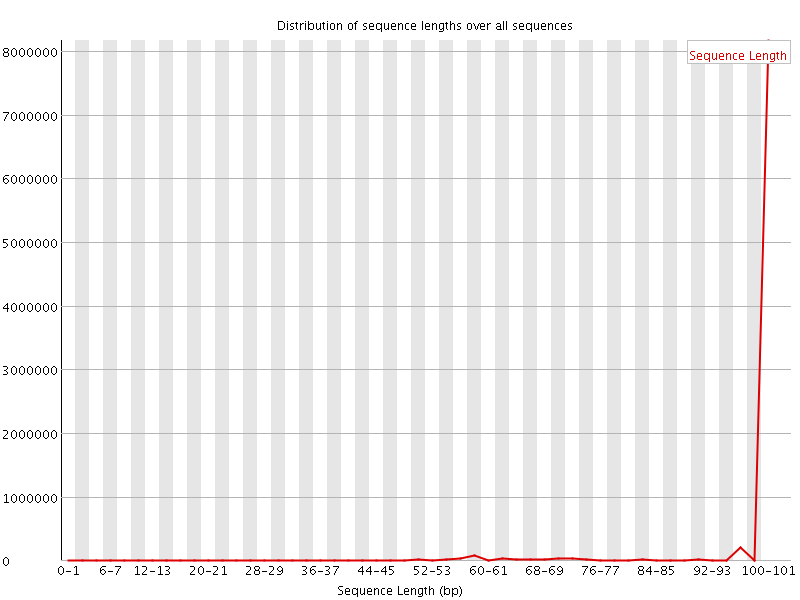
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels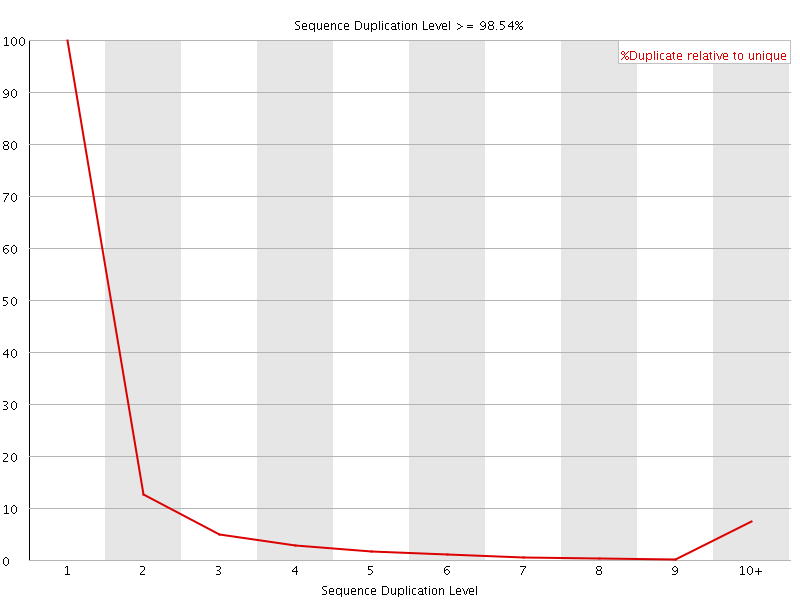
![[WARN]](Icons/warning.png) Overrepresented sequences
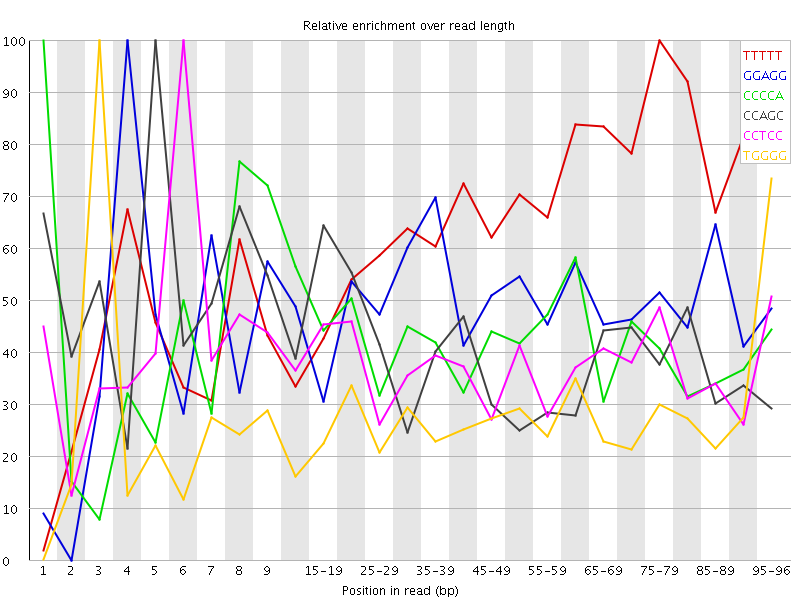
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content