![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-7.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 6722214 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 46 |
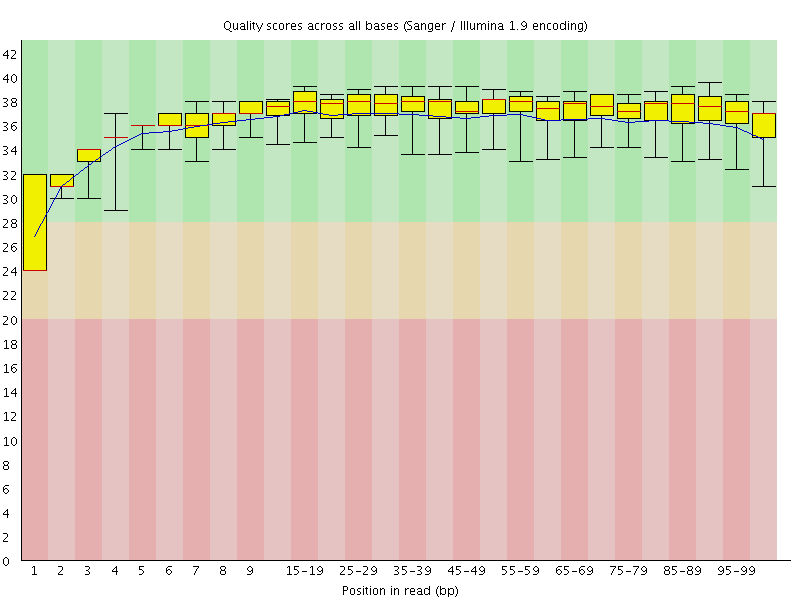
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
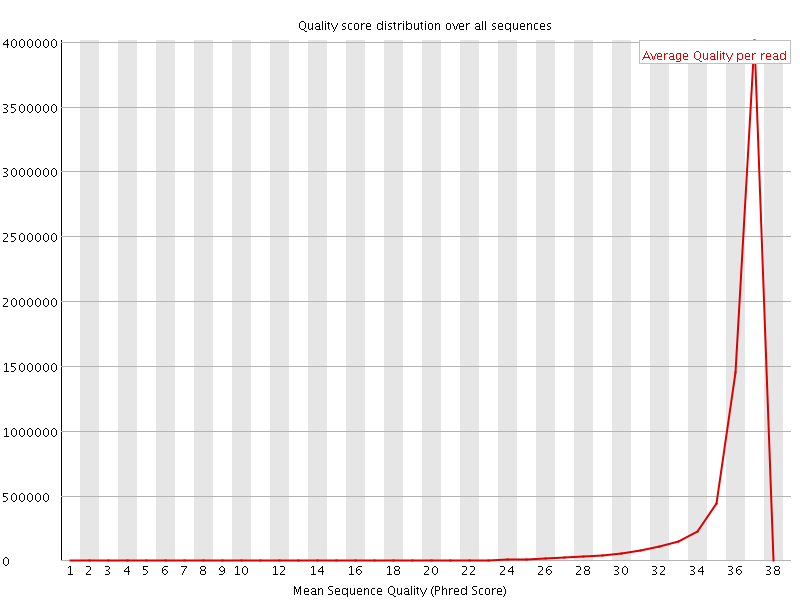
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
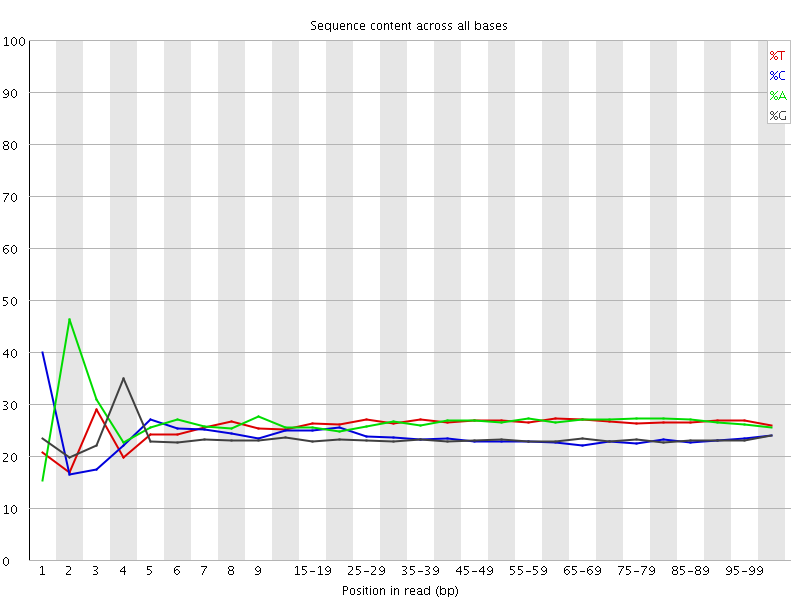
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
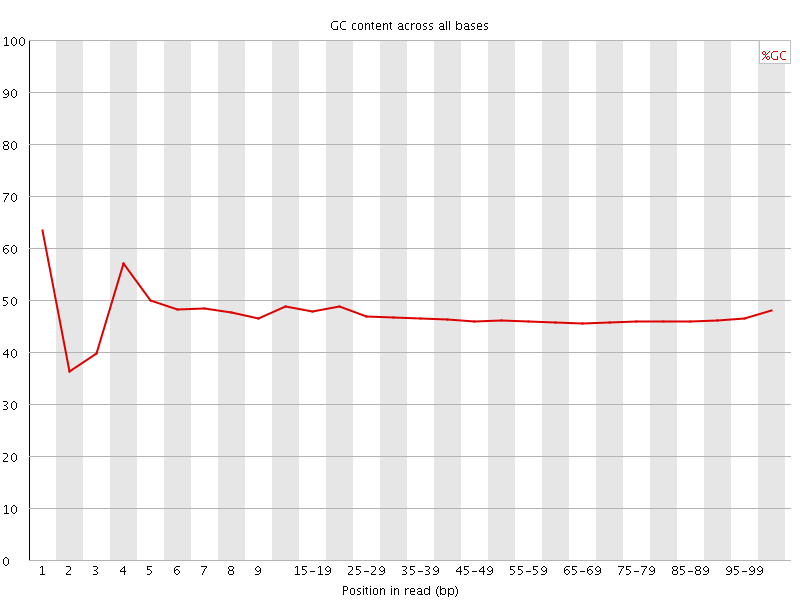
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
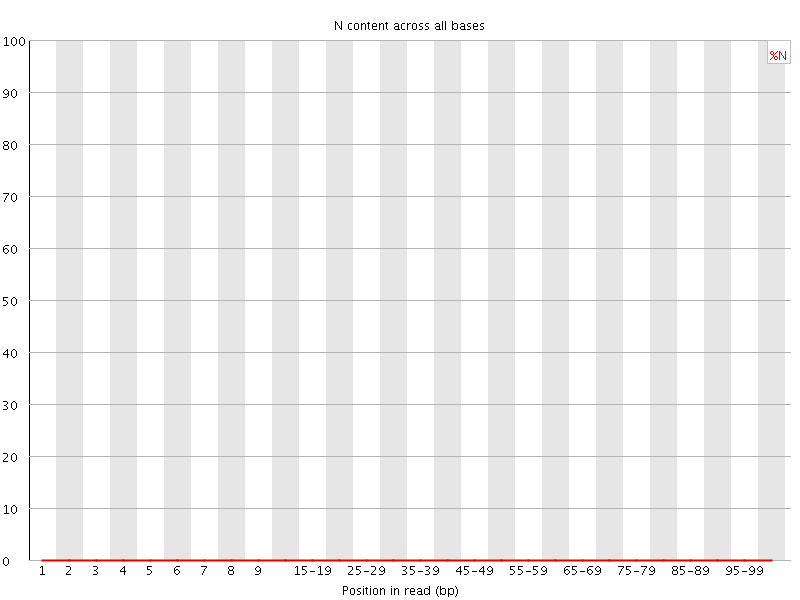
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
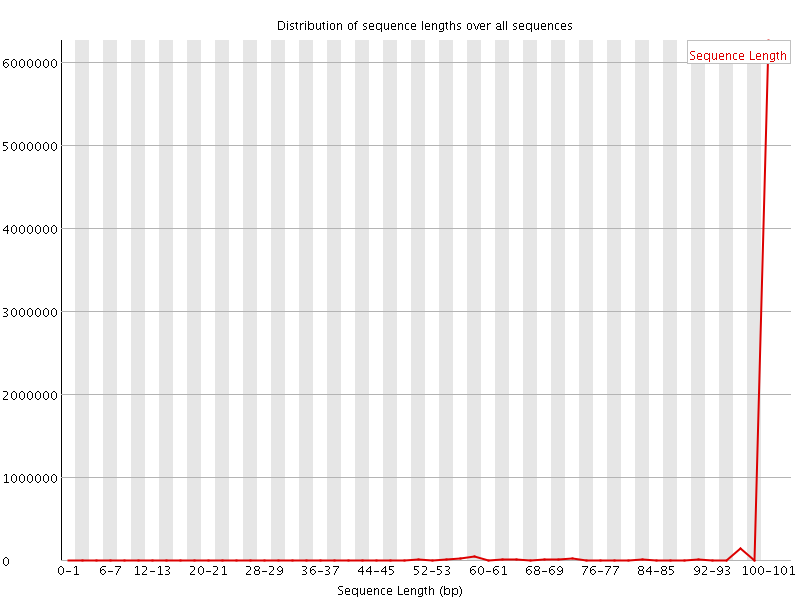
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 9994 | 0.14867125622599936 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 9919 | 0.14755555238199794 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 9814 | 0.145993567000396 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 9795 | 0.1457109220265823 | No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 9368 | 0.13935884814140104 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 9101 | 0.13538694245675606 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 8760 | 0.13031420897936305 | No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA | 8753 | 0.1302100766205896 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 8738 | 0.1299869358517893 | No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC | 8649 | 0.128662967290241 | No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT | 8497 | 0.12640180749973148 | No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT | 8421 | 0.12527122760447673 | No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA | 7991 | 0.1188745255655354 | No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC | 7957 | 0.11836873982292145 | No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA | 7595 | 0.11298360926920803 | No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC | 7557 | 0.11241831932158065 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 7378 | 0.10975550614723067 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 7371 | 0.1096513737884572 | No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT | 7229 | 0.10753897451048122 | No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG | 7199 | 0.10709269297288065 | No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA | 7184 | 0.10686955220408037 | No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC | 7157 | 0.10646789882023987 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 6967 | 0.10364144908210302 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 6948 | 0.10335880410828932 | No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA | 6776 | 0.10080012329271278 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
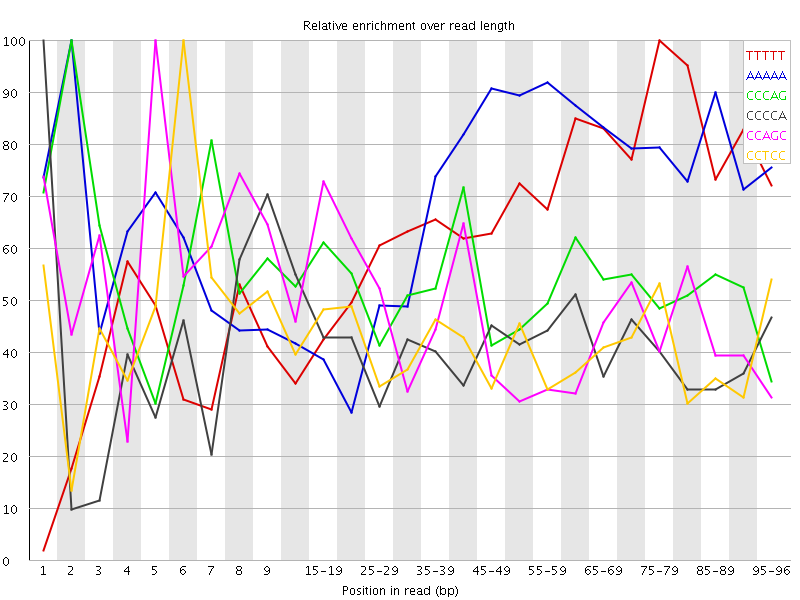
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 2641855 | 3.2269456 | 4.895105 | 75-79 |
| AAAAA | 2617955 | 3.040673 | 4.365095 | 2 |
| CCCAG | 1505905 | 2.8996425 | 5.428796 | 2 |
| CCCCA | 1322045 | 2.505746 | 6.088839 | 1 |
| CCAGC | 1251825 | 2.4104078 | 5.0971494 | 5 |
| CCTCC | 1222765 | 2.34104 | 5.6710687 | 6 |
| TGGGG | 1143535 | 2.33206 | 8.288691 | 3 |
| GGGGG | 996230 | 2.3094144 | 5.907885 | 4 |
| CTGCC | 1177185 | 2.2896373 | 5.5039673 | 7 |
| GGAAG | 1196775 | 2.1042671 | 5.098763 | 4 |
| CCACC | 1106305 | 2.0968418 | 5.95184 | 1 |
| TGGCT | 1127115 | 1.9904466 | 6.492927 | 3 |
| CCCTG | 1005085 | 1.9549006 | 5.244023 | 1 |
| TGCAG | 1101450 | 1.9256265 | 5.4602847 | 3 |
| GCCCA | 939680 | 1.8093681 | 5.7888975 | 2 |
| GGCTC | 909970 | 1.798064 | 7.8647113 | 1 |
| TGGCA | 1007905 | 1.762085 | 5.1416163 | 3 |
| GCTCC | 843365 | 1.6403539 | 7.627665 | 2 |
| GATGG | 888115 | 1.5773664 | 5.041971 | 1 |
| TGCCC | 810240 | 1.5759254 | 9.713757 | 3 |
| TAAAA | 1337875 | 1.5696329 | 5.889064 | 1 |
| TGAGG | 829700 | 1.4736165 | 5.003374 | 1 |
| CATGG | 837375 | 1.4639533 | 36.809944 | 1 |
| ATGGA | 939875 | 1.4538063 | 10.389134 | 2 |
| GGCCC | 630030 | 1.3929487 | 5.5849447 | 1 |
| TGGGT | 739155 | 1.3260928 | 5.7498965 | 3 |
| ATGGG | 746455 | 1.3257664 | 11.860283 | 2 |
| ATGTG | 836415 | 1.3068727 | 9.802917 | 2 |
| ATGGC | 730525 | 1.2771512 | 11.4350815 | 2 |
| TGAGA | 789200 | 1.2207409 | 5.0232034 | 1 |
| ATGAA | 877500 | 1.1821065 | 7.8245893 | 2 |
| ATGCC | 671630 | 1.1557962 | 9.312797 | 2 |
| ATGTT | 811710 | 1.1157334 | 6.235039 | 2 |
| CATGA | 705725 | 1.0745232 | 22.989094 | 1 |
| CATGT | 685375 | 1.0541042 | 27.77104 | 1 |
| ATGCT | 685205 | 1.0538429 | 8.114627 | 2 |
| ATGAG | 677385 | 1.0477846 | 5.469965 | 2 |
| ATGGT | 626415 | 0.97875416 | 6.335554 | 2 |
| CATGC | 560170 | 0.9639865 | 28.131937 | 1 |
| ATGAC | 606080 | 0.9228057 | 6.303243 | 2 |
| ATGTA | 629365 | 0.85642064 | 7.0857334 | 2 |
| TAATG | 596425 | 0.81159675 | 5.2849865 | 1 |
| ATGCA | 522010 | 0.79480237 | 9.579981 | 2 |
| ATGTC | 493235 | 0.75859374 | 5.151902 | 2 |
| TAAGT | 548785 | 0.7467698 | 6.1612115 | 1 |