![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-8.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 8250800 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 46 |
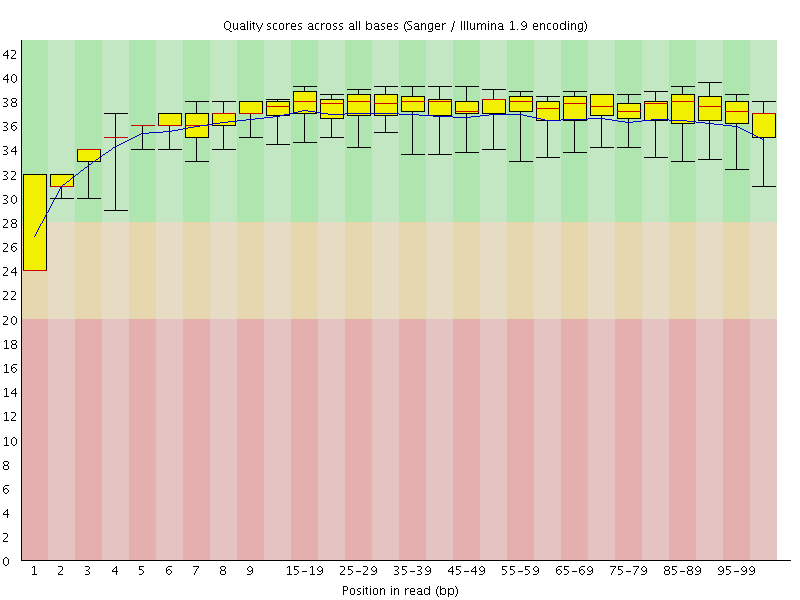
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
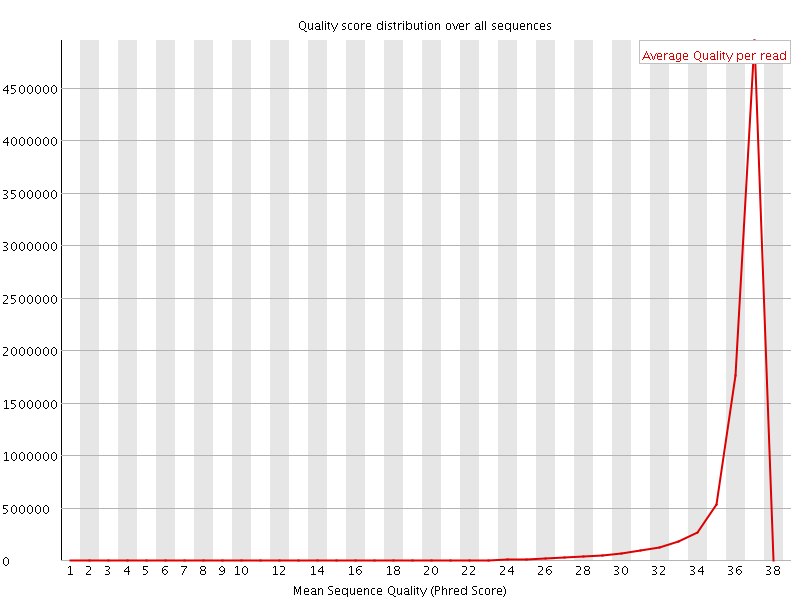
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
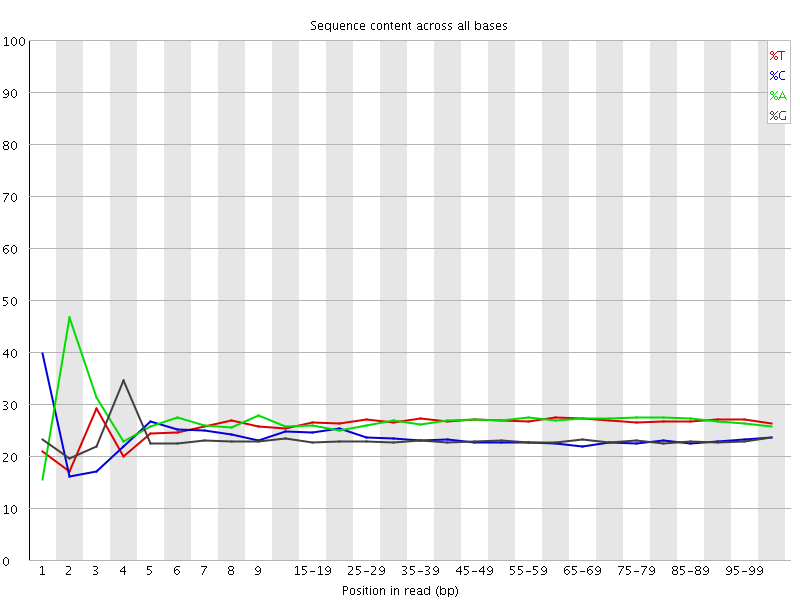
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
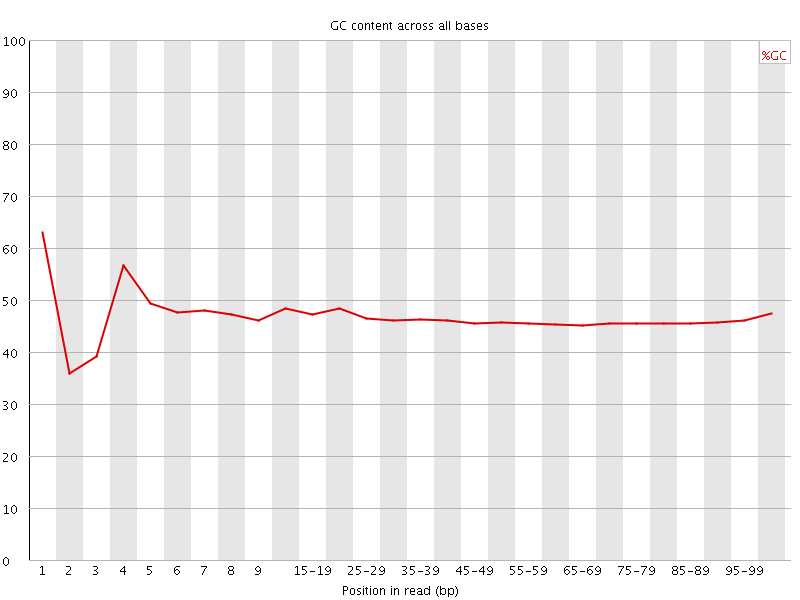
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
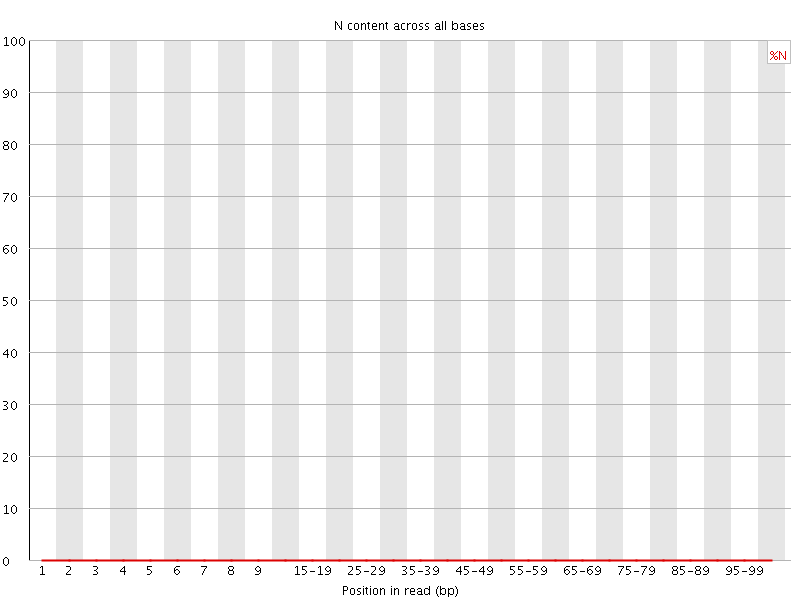
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
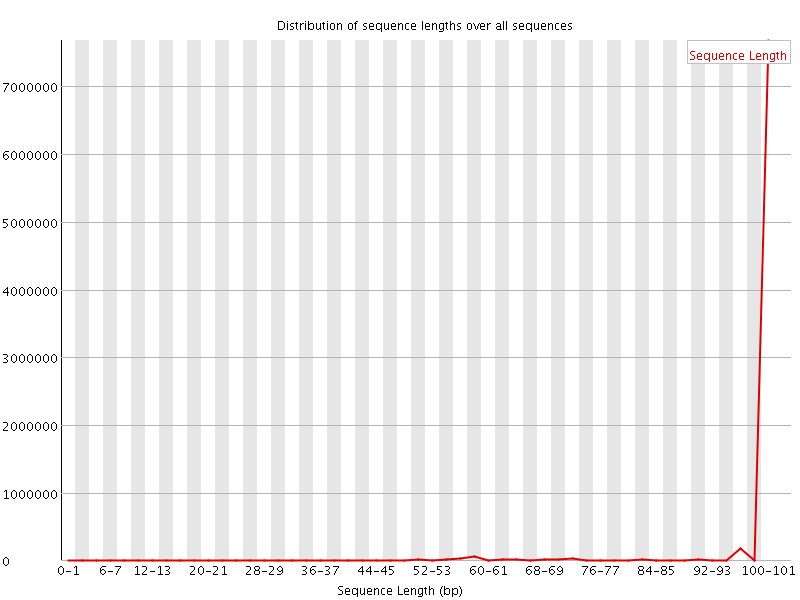
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 11851 | 0.14363455664905223 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 11836 | 0.14345275609637853 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 11758 | 0.1425073932224754 | No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 11538 | 0.13984098511659476 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 11450 | 0.1387744218742425 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 11410 | 0.138289620400446 | No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA | 10886 | 0.13193872109371213 | No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC | 10701 | 0.12969651427740342 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 10125 | 0.12271537305473408 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 10102 | 0.12243661220730112 | No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT | 9678 | 0.11729771658505841 | No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT | 9659 | 0.11706743588500508 | No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA | 9253 | 0.11214670092597082 | No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC | 9197 | 0.11146797886265575 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 8999 | 0.10906821156736317 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 8973 | 0.10875309060939546 | No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA | 8745 | 0.10598972220875551 | No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC | 8708 | 0.10554128084549377 | No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG | 8527 | 0.10334755417656469 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 8516 | 0.10321423377127065 | No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT | 8508 | 0.10311727347651137 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 8488 | 0.10287487273961313 | No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA | 8367 | 0.10140834828137876 | No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA | 8284 | 0.10040238522325108 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
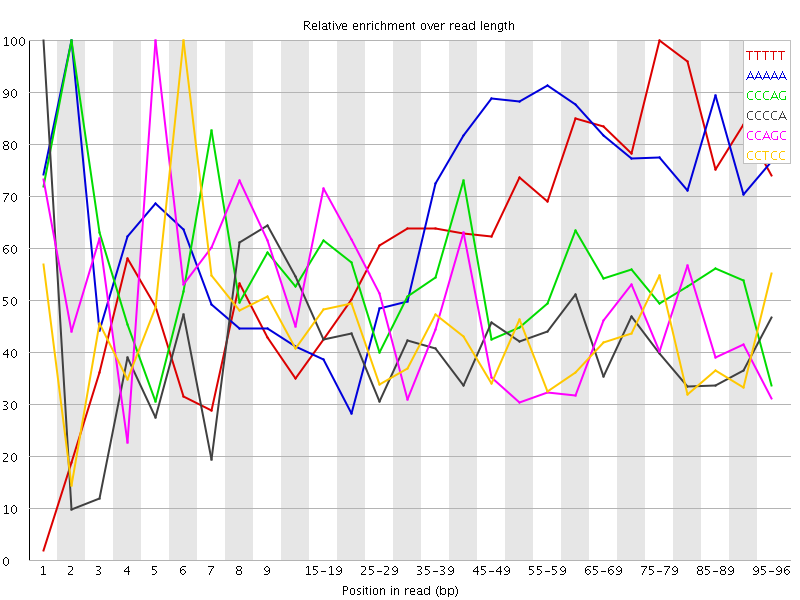
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 3306405 | 3.1727278 | 4.776519 | 75-79 |
| AAAAA | 3291015 | 3.0051918 | 4.3581595 | 2 |
| CCCAG | 1796275 | 2.8945365 | 5.348742 | 2 |
| CCCCA | 1581130 | 2.5082693 | 6.0659814 | 1 |
| CCAGC | 1504395 | 2.424198 | 5.172347 | 5 |
| CCTCC | 1468400 | 2.3526528 | 5.604707 | 6 |
| TGGGG | 1374205 | 2.3440309 | 8.151389 | 3 |
| GGGGG | 1190025 | 2.3438332 | 5.7783737 | 4 |
| CTGCC | 1404415 | 2.2856436 | 5.5608697 | 7 |
| CCACC | 1333830 | 2.1159582 | 5.903433 | 1 |
| GGAAG | 1452345 | 2.103338 | 5.020593 | 4 |
| TGGCT | 1368410 | 1.9900762 | 6.251745 | 3 |
| CCCTG | 1203880 | 1.959279 | 5.227167 | 1 |
| TGCAG | 1327710 | 1.9118322 | 5.6273212 | 3 |
| GCCCA | 1125525 | 1.8136828 | 5.614962 | 2 |
| GGCTC | 1083195 | 1.7906854 | 7.3626328 | 1 |
| TGGCA | 1235490 | 1.7790406 | 5.104687 | 3 |
| GCTCC | 1002940 | 1.6322551 | 7.0667596 | 2 |
| TGCCC | 962470 | 1.5663913 | 9.576345 | 3 |
| TAAAA | 1677330 | 1.5469198 | 5.920756 | 1 |
| TGAGG | 1011820 | 1.479958 | 5.042492 | 1 |
| CATGG | 1021135 | 1.4703807 | 36.164043 | 1 |
| ATGGA | 1157425 | 1.4516885 | 10.046238 | 2 |
| GGCCC | 746105 | 1.4020767 | 5.550292 | 1 |
| TGGCC | 824355 | 1.3627837 | 5.163738 | 3 |
| TGGGT | 897415 | 1.3257035 | 5.609238 | 3 |
| ATGGG | 898485 | 1.3141867 | 11.603185 | 2 |
| ATGTG | 1025655 | 1.299238 | 9.604631 | 2 |
| ATGGC | 891535 | 1.2837636 | 11.115032 | 2 |
| ATGAA | 1099660 | 1.1826966 | 7.850102 | 2 |
| ATGCC | 813715 | 1.1535048 | 9.081499 | 2 |
| TGAGC | 784950 | 1.1302866 | 5.0294313 | 1 |
| ATGTT | 1014420 | 1.1128753 | 6.23479 | 2 |
| CATGA | 873690 | 1.0787935 | 23.177553 | 1 |
| ATGCT | 854860 | 1.066063 | 8.10213 | 2 |
| CATGT | 846415 | 1.0555315 | 27.55346 | 1 |
| ATGAG | 833575 | 1.0455029 | 5.3744917 | 2 |
| ATGGT | 778065 | 0.9856061 | 6.269948 | 2 |
| CATGC | 688955 | 0.9766476 | 28.488892 | 1 |
| ATGAC | 750860 | 0.9271285 | 6.2317 | 2 |
| ATGTA | 794520 | 0.8630314 | 7.0023484 | 2 |
| TAATG | 751125 | 0.81589454 | 5.0900874 | 1 |
| ATGCA | 654045 | 0.80758566 | 9.71299 | 2 |
| TAAGT | 687815 | 0.7471253 | 6.1564207 | 1 |