![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-7-9.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 9230846 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 45 |
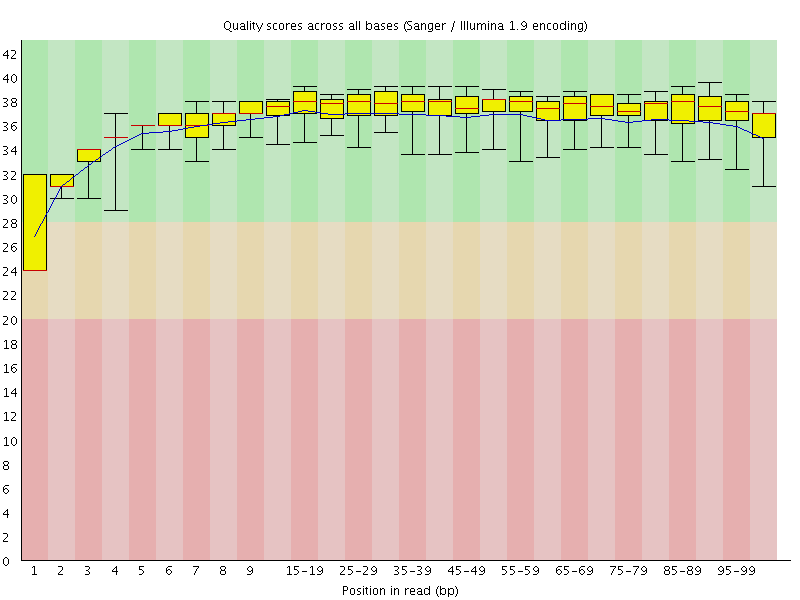
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
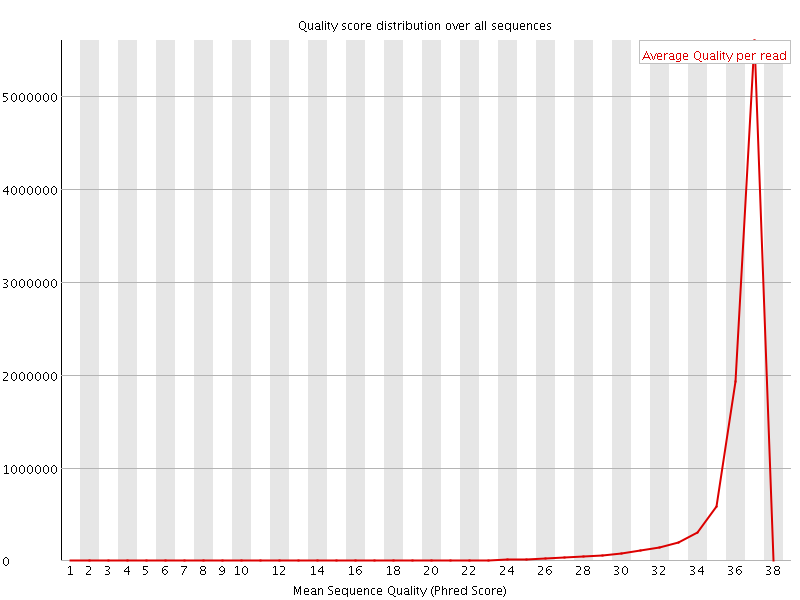
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
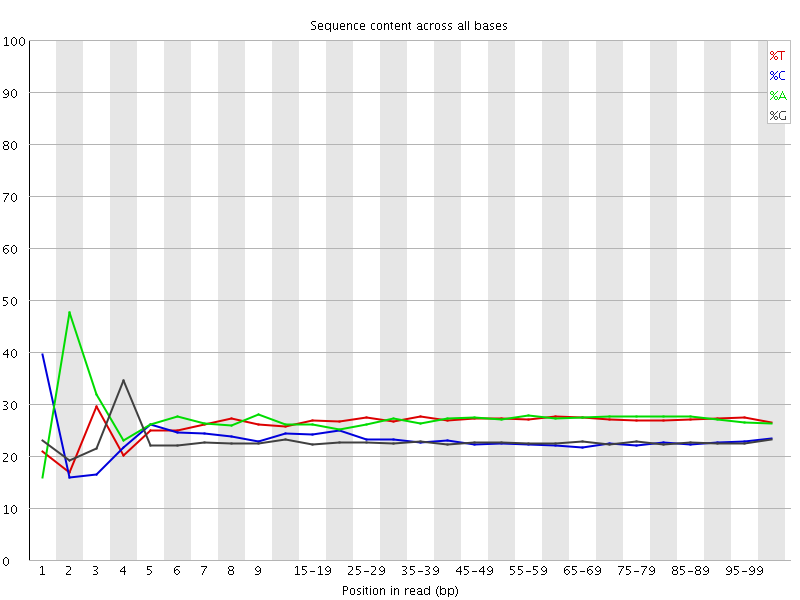
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
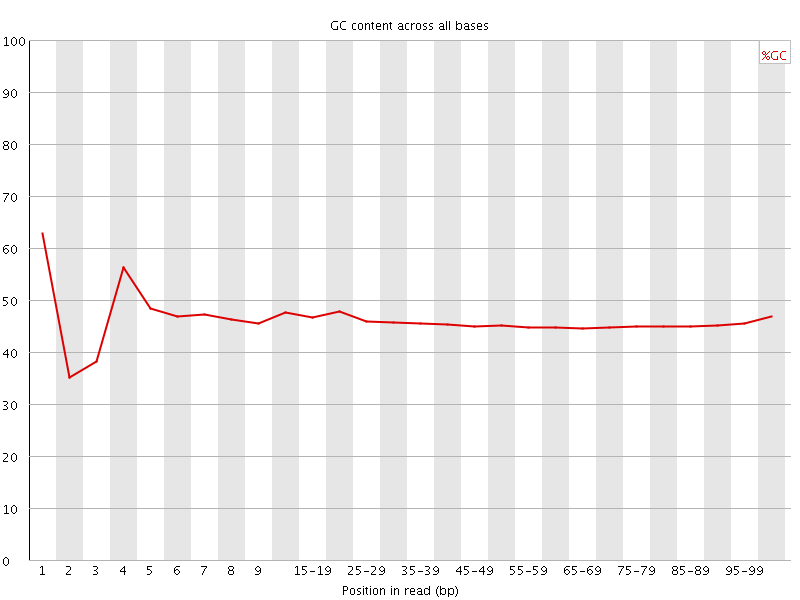
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
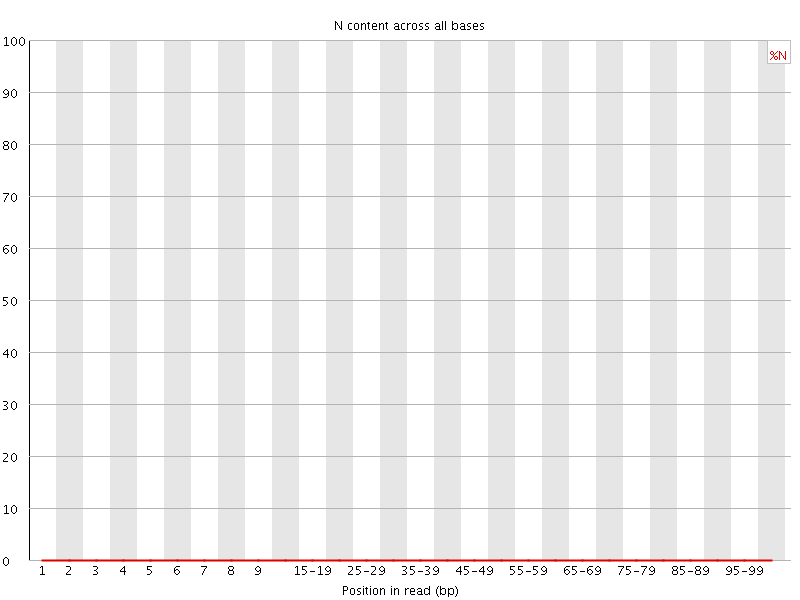
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
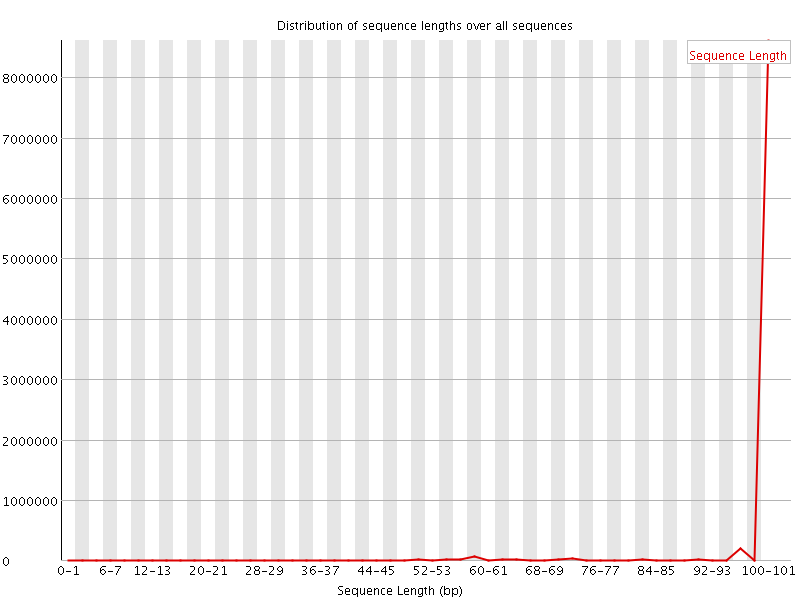
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 13421 | 0.14539295748190362 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 13271 | 0.14376797099637453 | No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG | 12012 | 0.1301289177611673 | No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC | 11711 | 0.1268681115468723 | No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG | 11662 | 0.12633728262826616 | No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG | 11582 | 0.1254706231693173 | No Hit |
| GAGCTACCATCTGATCTGTCTGTCTTGACCACCCGGAGTCCCACTGTCCCCAGCCAGAATCCCAGTAGACTA | 11462 | 0.12417063398089405 | No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA | 11455 | 0.12409480127823604 | No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC | 11438 | 0.12391063614320941 | No Hit |
| TAGTCTACTGGGATTCTGGCTGGGGACAGTGGGACTCCGGGTGGTCAAGACAGACAGATCAGATGGTAGCTC | 11284 | 0.1222423166847329 | No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT | 10906 | 0.11814735074119967 | No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT | 10849 | 0.11752985587669862 | No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC | 10784 | 0.11682569506630269 | No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA | 10775 | 0.11672819587717097 | No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA | 10208 | 0.1105857469618711 | No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG | 10179 | 0.11027158290800214 | No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA | 10012 | 0.10846243128744645 | No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC | 10011 | 0.1084515980442096 | No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT | 9936 | 0.10763910480144506 | No Hit |
| ATTCAGCTGCCAAGTAACTTAGCACACCCATTCTTATAAGATAATGGCAA | 9928 | 0.10755243885555019 | No Hit |
| TAATGAGCATGAAGTCACCACACGGAGGAAAATGTAAATGTGTAAACCTC | 9907 | 0.1073249407475761 | No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT | 9904 | 0.10729244101786554 | No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG | 9283 | 0.10056499696777521 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
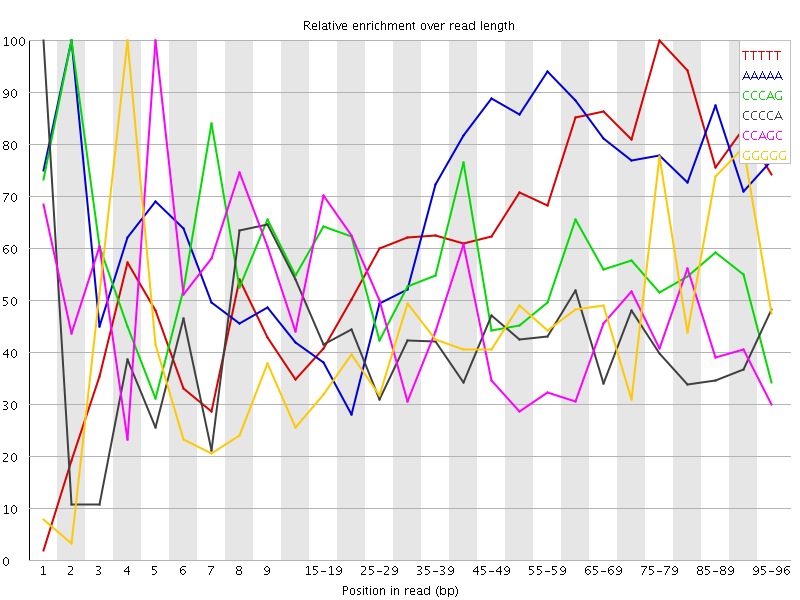
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 3940115 | 3.1828754 | 4.8183336 | 75-79 |
| AAAAA | 3923060 | 3.0239356 | 4.3675714 | 2 |
| CCCAG | 1934560 | 2.9060853 | 5.190659 | 2 |
| CCCCA | 1700795 | 2.5168054 | 6.045392 | 1 |
| CCAGC | 1603975 | 2.4094822 | 5.225475 | 5 |
| GGGGG | 1267445 | 2.3800256 | 5.2077036 | 4 |
| CCTCC | 1573510 | 2.3503892 | 5.716723 | 6 |
| TGGGG | 1481235 | 2.3496766 | 7.328133 | 3 |
| CTGCC | 1528645 | 2.3179572 | 5.587548 | 7 |
| CCACC | 1451325 | 2.1476443 | 6.002725 | 1 |
| TGGCT | 1522485 | 2.009742 | 6.4317846 | 3 |
| TGCAG | 1453905 | 1.9013004 | 6.117821 | 3 |
| GGCTC | 1179305 | 1.8153207 | 6.5730896 | 1 |
| TGGCA | 1359150 | 1.7773875 | 5.236168 | 3 |
| GCTCC | 1084485 | 1.644456 | 6.226146 | 2 |
| GATGG | 1184555 | 1.572528 | 5.109635 | 1 |
| TGCCC | 1030805 | 1.5630587 | 9.4135475 | 3 |
| TAAAA | 1971415 | 1.5339043 | 5.76929 | 1 |
| CATGG | 1116810 | 1.4604745 | 38.046883 | 1 |
| ATGGA | 1311650 | 1.4572035 | 10.540634 | 2 |
| AGACA | 1242600 | 1.3472012 | 5.014646 | 2 |
| TGGCC | 869095 | 1.33781 | 5.14678 | 3 |
| TGGGT | 978445 | 1.3111495 | 5.830887 | 3 |
| ATGTG | 1159140 | 1.2999022 | 9.776243 | 2 |
| ATGGG | 976580 | 1.2964358 | 11.882715 | 2 |
| ATGGC | 975965 | 1.276289 | 11.385974 | 2 |
| ATGAA | 1275160 | 1.1855667 | 7.990884 | 2 |
| ATGCC | 898035 | 1.1568567 | 8.9615555 | 2 |
| TGAGC | 868230 | 1.1354016 | 5.1916394 | 1 |
| ATGTT | 1154195 | 1.0934166 | 6.596095 | 2 |
| CATGA | 977100 | 1.0693325 | 24.295666 | 1 |
| ATGCT | 965470 | 1.0665597 | 7.8415856 | 2 |
| CATGT | 956925 | 1.0571198 | 29.323277 | 1 |
| ATGAG | 925025 | 1.0276748 | 5.573728 | 2 |
| ATGGT | 885140 | 0.99262875 | 6.649436 | 2 |
| CATGC | 748230 | 0.9638764 | 29.084887 | 1 |
| ATGAC | 864070 | 0.9456332 | 6.58165 | 2 |
| ATGTA | 930730 | 0.8734892 | 7.4471283 | 2 |
| ATGCA | 741275 | 0.811247 | 10.055528 | 2 |
| ATGTC | 689875 | 0.76210845 | 5.131507 | 2 |
| TAAGT | 799800 | 0.75061154 | 5.860905 | 1 |