| Sequence |
Count |
Percentage |
Possible Source |
| TGAGACCGCACCAGCGACACA |
20136 |
0.36488718324708275 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
19862 |
0.35992199213615206 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
11930 |
0.21618514581534057 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
11878 |
0.21524284677239022 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11241 |
0.2036996834962484 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11131 |
0.20170635859769956 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
10696 |
0.19382366468071102 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
10624 |
0.19251894292893362 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
10404 |
0.18853229313183598 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
10254 |
0.18581412281563303 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
10085 |
0.1827516509260444 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
10063 |
0.18235298594633462 |
No Hit |
| * |
9842 |
0.1783482150137956 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
9792 |
0.177442158241728 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
9737 |
0.17644549579245355 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
9374 |
0.16986752362724244 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
9194 |
0.1666057192477989 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
8905 |
0.16136871110524792 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
8889 |
0.16107877293818626 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
8815 |
0.15973780891552616 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
8629 |
0.1563672777234345 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
8484 |
0.15373971308443835 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
8386 |
0.15196384181118575 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
8327 |
0.15089469482014592 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
7822 |
0.14174352142226268 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
7812 |
0.14156231006784917 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
7777 |
0.1409280703274018 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
7728 |
0.1400401346907755 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
7718 |
0.139858923336362 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
7508 |
0.13605348489367786 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
7472 |
0.13540112401778914 |
No Hit |
| CATGTACAAGGCCTTGCTACCTCAGCAGTCCTACAGCTTGGCCCAGCCGC |
7091 |
0.12849697141463368 |
No Hit |
| GGTGATGCCAAGGACGATGGGATGTGGGGACCGACGTAGTGAGGTGGCGG |
6884 |
0.12474589637827363 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
6794 |
0.12311499418855186 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
6777 |
0.12280693488604887 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6580 |
0.11923707120410232 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
6510 |
0.11796859172320763 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
6492 |
0.11764241128526327 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
6484 |
0.11749744220173246 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
6446 |
0.11680883905496105 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
6424 |
0.11641017407525128 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6422 |
0.11637393180436859 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
6409 |
0.11613835704363097 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
6197 |
0.11229667633006417 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
6157 |
0.11157183091241003 |
No Hit |
| GAGACCGCACCAGCGACACC |
6060 |
0.1098140807745988 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6018 |
0.10905299308606199 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
5905 |
0.1070053047811891 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
5873 |
0.10642542844706582 |
No Hit |
| GGTGTCGCTGGTGCGGTCTC |
5866 |
0.10629858049897635 |
No Hit |
| CGAGACCGCGCCCCAGGCCCAGCACTCACAAATTCCTGGTCGTGGTTATT |
5827 |
0.10559185621676356 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
5813 |
0.10533816032058463 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
5773 |
0.10461331490293051 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
5669 |
0.1027287168170298 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
5648 |
0.1023481729727614 |
No Hit |
| GGCACAAGGGTACCTACGGGCTGCTGCGGCGGCGAGAGGACTGGCCCTCC |
5647 |
0.10233005183732005 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
5619 |
0.10182266004496215 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5531 |
0.10022800012612311 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
5521 |
0.10004678877170957 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality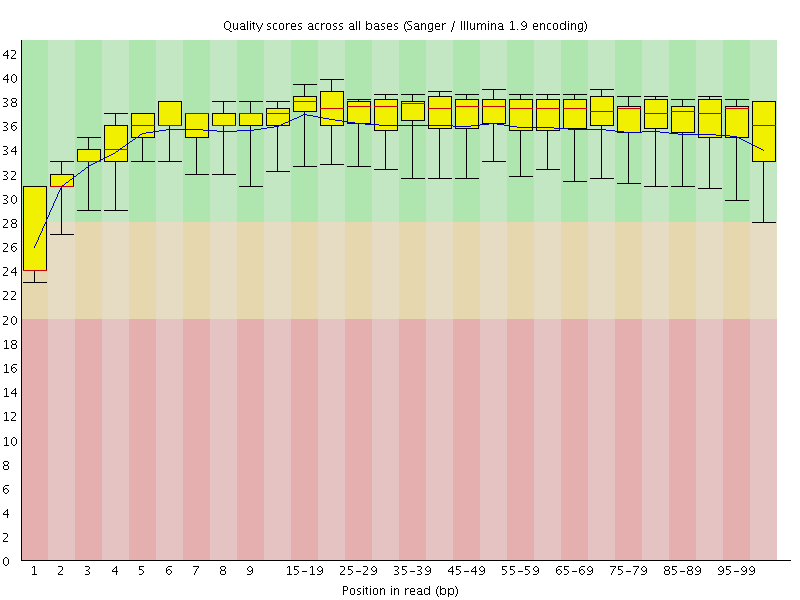
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores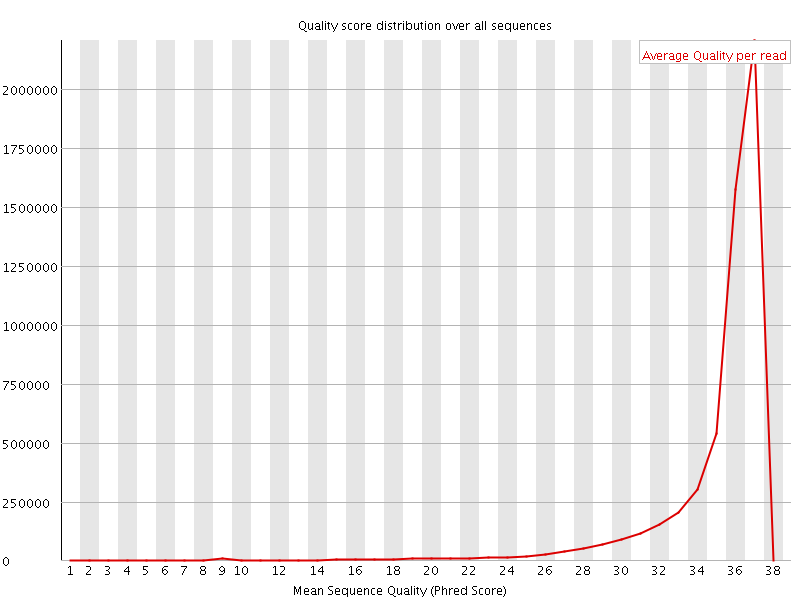
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content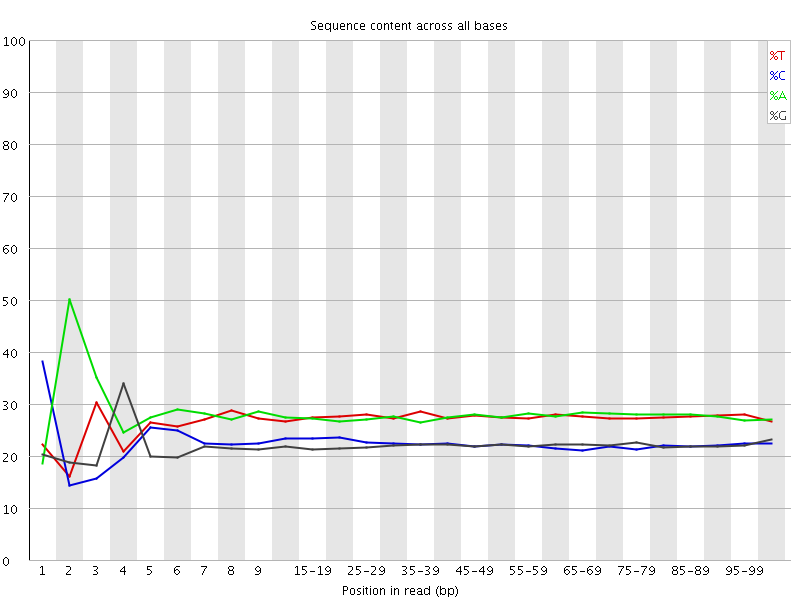
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution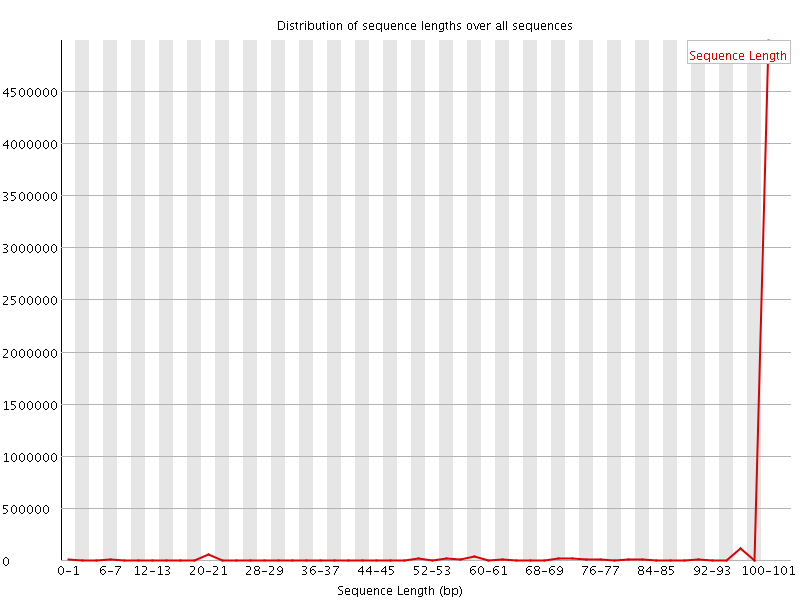
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels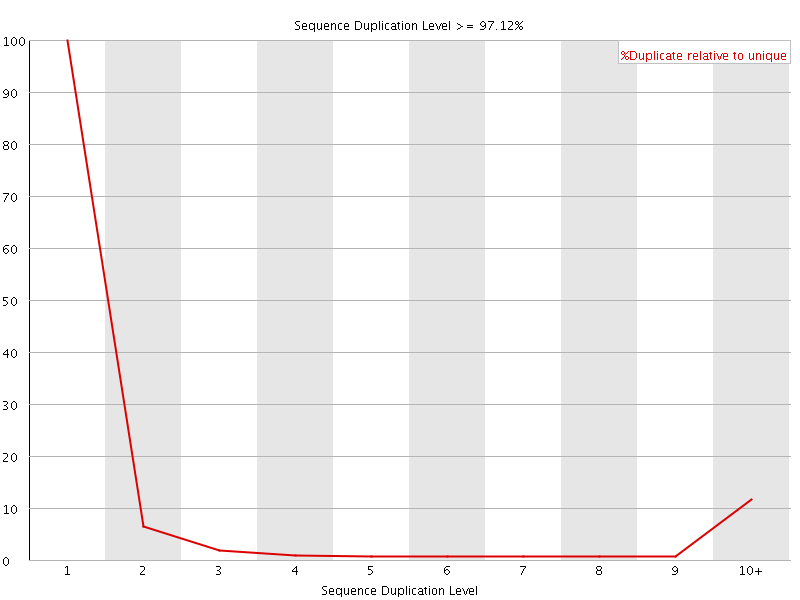
![[WARN]](Icons/warning.png) Overrepresented sequences
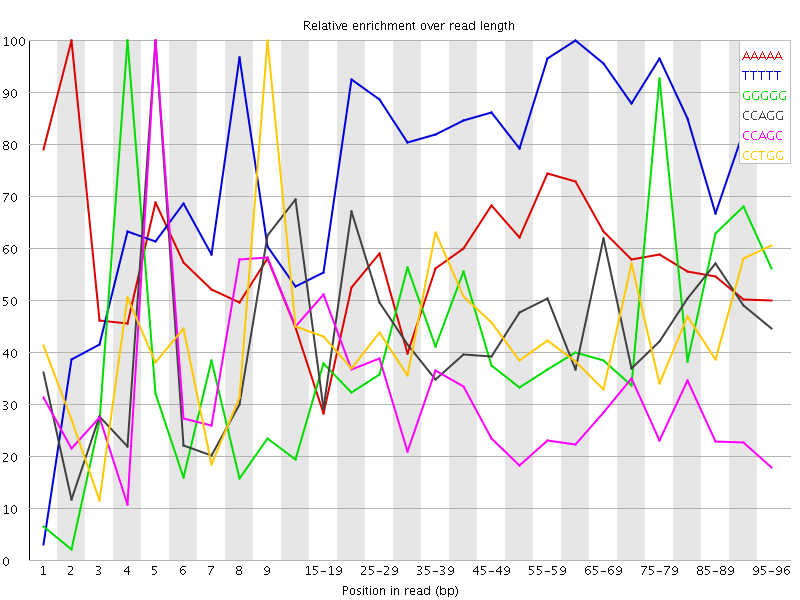
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content