| Sequence |
Count |
Percentage |
Possible Source |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10770 |
0.22932302650182096 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10622 |
0.2261716980039315 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
10606 |
0.2258310138419975 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
10559 |
0.2248302541163164 |
No Hit |
| * |
10172 |
0.21658995594953787 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
9262 |
0.19721354423954182 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
9213 |
0.196170198993619 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8646 |
0.184097204005083 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8631 |
0.1837778126032699 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
8612 |
0.18337325016097328 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
8611 |
0.1833519574008524 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
8473 |
0.18041355650417168 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
8458 |
0.18009416510235857 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
8399 |
0.17883789225522695 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
8218 |
0.1749839026733486 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
7642 |
0.16271927284372478 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
7635 |
0.16257022352287864 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
7480 |
0.15926984570414304 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
7399 |
0.15754513213435223 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7398 |
0.15752383937423134 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7394 |
0.15743866833374784 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
7377 |
0.15707669141169298 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7338 |
0.15624627376697883 |
No Hit |
| TGAGACCGCACCAGCGACACA |
6998 |
0.14900673532588143 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6964 |
0.14828278148177168 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6907 |
0.14706909415488184 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
6887 |
0.14664323895246434 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6883 |
0.14655806791198084 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
6805 |
0.1448972326225526 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
6791 |
0.14459913398086036 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
6745 |
0.14361966701530013 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
6734 |
0.14338544665397052 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
6463 |
0.13761510866121346 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6393 |
0.13612461545275223 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
6361 |
0.13544324712888423 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6335 |
0.13488963536574147 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
5844 |
0.12443489014639199 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
5789 |
0.12326378833974387 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5616 |
0.11958014083883253 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
5571 |
0.1186219666333932 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
5522 |
0.11757862138747031 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
5346 |
0.11383109560619635 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
5318 |
0.11323489832281187 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
5268 |
0.11217026031676815 |
No Hit |
| GCACGTGGGAGAGTAAGCAGGCCAGGTAGAGCTGCAGAAAACTAAGGCCC |
5267 |
0.11214896755664726 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
5236 |
0.11148889199290013 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
5234 |
0.1114463064726584 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
5220 |
0.11114820783096616 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
5219 |
0.11112691507084527 |
No Hit |
| TAACCAGAGTTAGTAGTGTATGTAAAATTCTGACTGTTCTTGCAACAATA |
5201 |
0.11074364538866951 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
5192 |
0.11055201054758165 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
5168 |
0.11004098430468065 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
5098 |
0.10855049109621943 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
5074 |
0.10803946485331843 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
5040 |
0.10731551100920868 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
5025 |
0.10699611960739558 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
5015 |
0.10678319200618683 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
5009 |
0.10665543544546158 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
4988 |
0.1062082874829232 |
No Hit |
| AATCTCTTTCCTTTATAAATTACCCAGTCTTGCATATGTCTTTATTAGCA |
4978 |
0.10599535988171445 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
4962 |
0.10565467571978046 |
No Hit |
| GTATCTTAATGATGTATATAATTGCCTTCAATCCCCTTCTCACCCCACCC |
4894 |
0.10420676803156098 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
4891 |
0.10414288975119837 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
4863 |
0.10354669246781387 |
No Hit |
| CATGACTATAAATATGAAGCAAATATGATTATCCTCACAAGTTCTGGAAT |
4858 |
0.10344022866720949 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
4856 |
0.10339764314696775 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
4851 |
0.10329117934636337 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
4837 |
0.10299308070467113 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
4828 |
0.10280144586358325 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
4827 |
0.10278015310346236 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
4792 |
0.10203490649923176 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
4779 |
0.10175810061766037 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
4761 |
0.10137483093548465 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
4743 |
0.10099156125330891 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
4735 |
0.1008212191723419 |
No Hit |
| CATGCCCACAGAGACTTGCACAACATGCAGAATGGCAGCACATTGGTAAG |
4701 |
0.10009726532823215 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality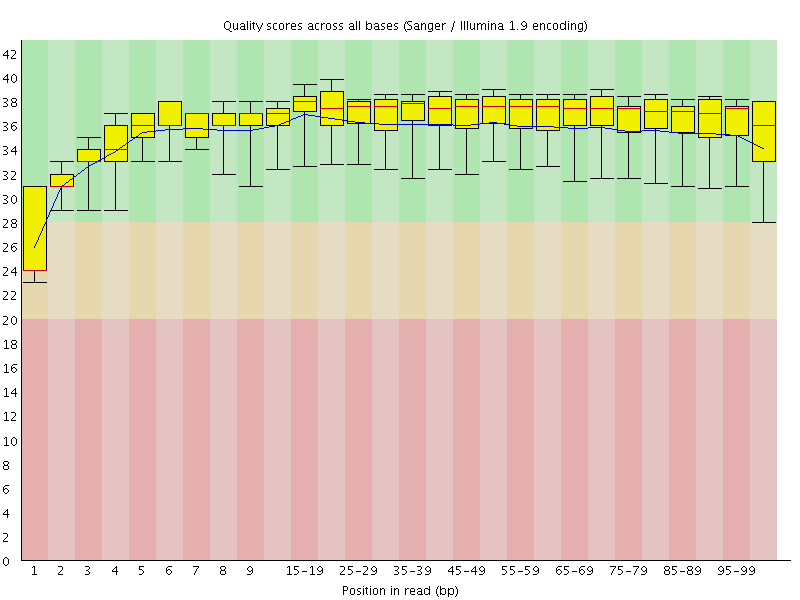
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores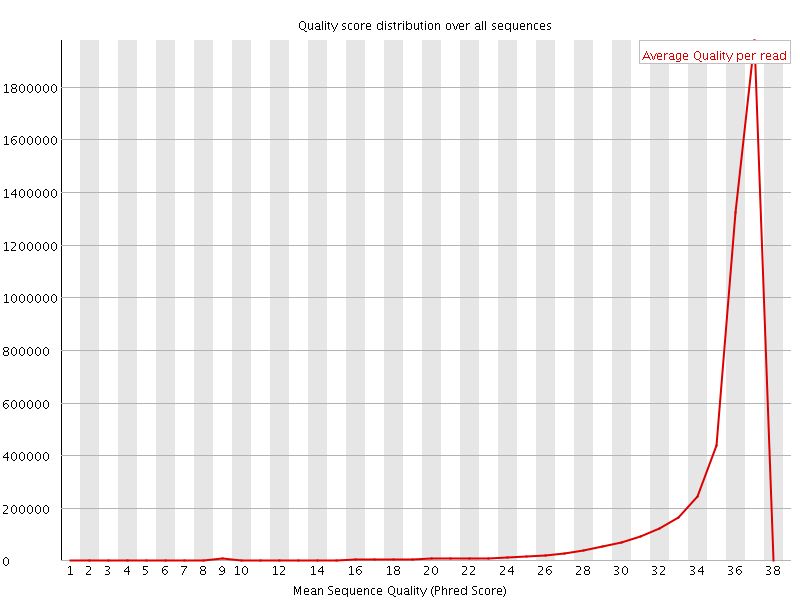
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content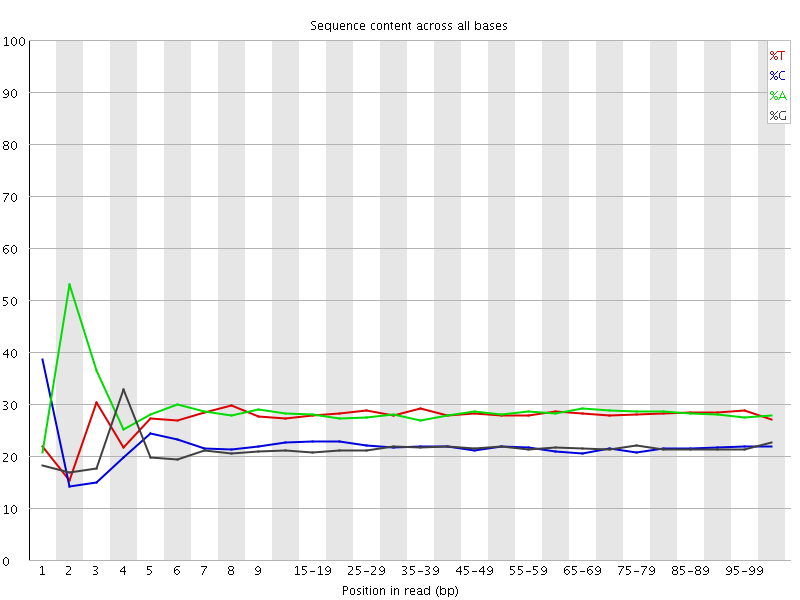
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content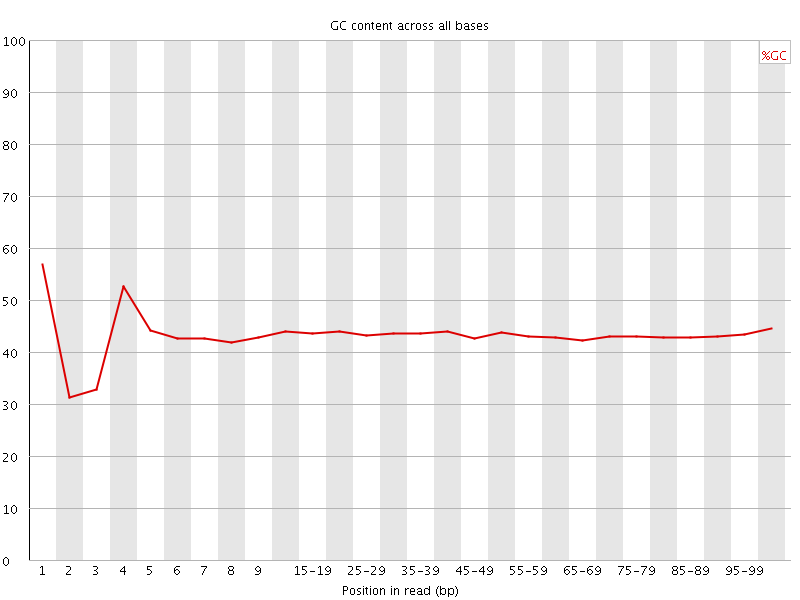
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content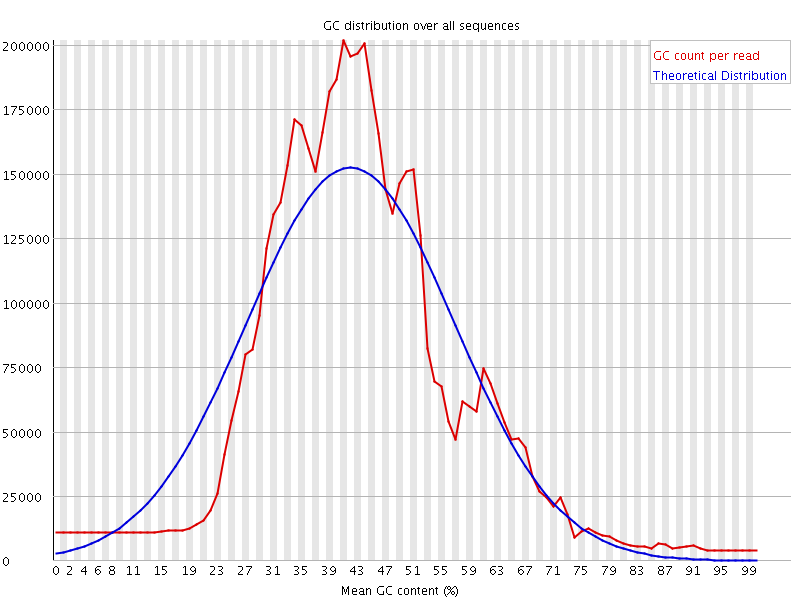
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution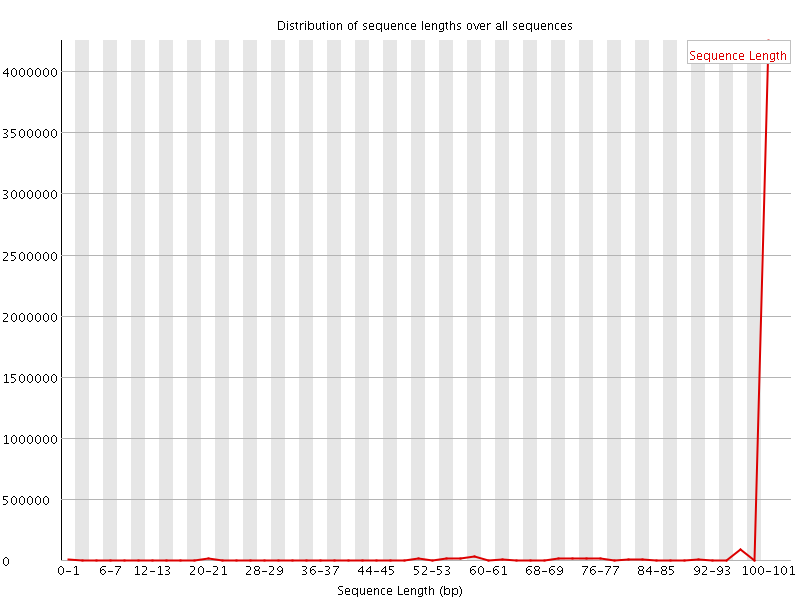
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels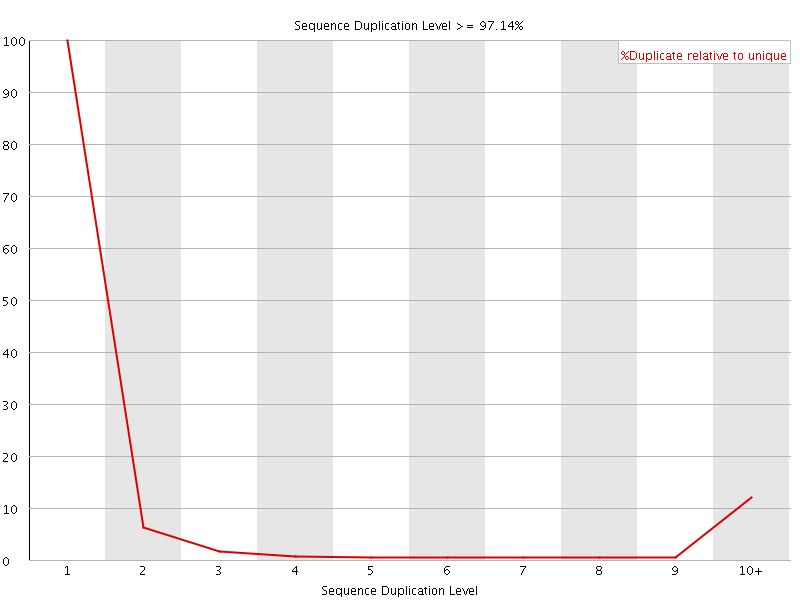
![[WARN]](Icons/warning.png) Overrepresented sequences
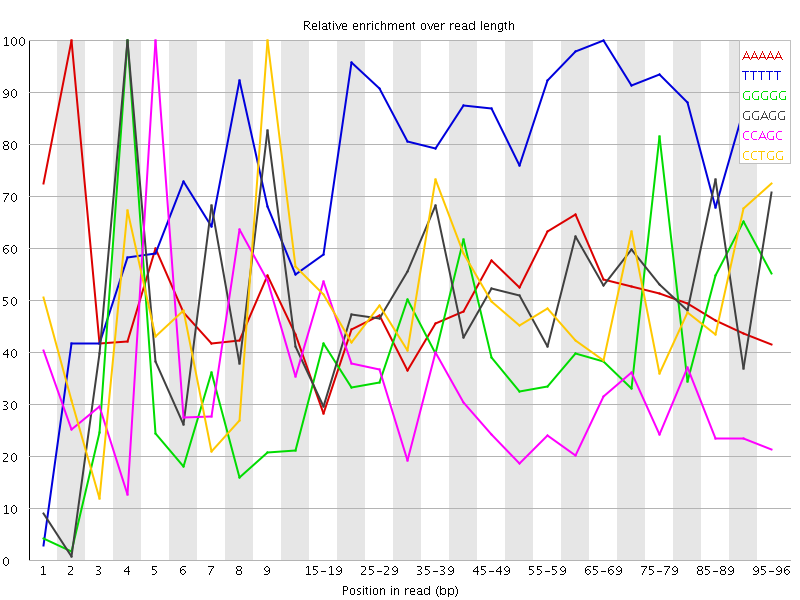
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content