| Sequence |
Count |
Percentage |
Possible Source |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
11875 |
0.2363881248161149 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
11826 |
0.2354127127642421 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
9858 |
0.19623697974208515 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
9820 |
0.19548053774267357 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
8944 |
0.1780425590193964 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
8907 |
0.17730602338839038 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8769 |
0.17455894454842202 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
8666 |
0.1725085886026486 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8662 |
0.1724289631290263 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
8429 |
0.16779077929052907 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
7898 |
0.15722049766717267 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
7825 |
0.15576733277356625 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7696 |
0.15319941124924805 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7672 |
0.15272165840751442 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
7660 |
0.1524827819866476 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
7621 |
0.15170643361883046 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
7472 |
0.1487403847264009 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
7412 |
0.14754600262206685 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
7372 |
0.14674974788584416 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
7362 |
0.1465506842017885 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
7016 |
0.1396630807334621 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
7000 |
0.139344578838973 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
6813 |
0.13562208794713188 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
6800 |
0.1353633051578595 |
No Hit |
| TGAGACCGCACCAGCGACACA |
6682 |
0.1330143536860025 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6628 |
0.13193940979210186 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
6623 |
0.13183987795007404 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6617 |
0.13172043973964065 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
6616 |
0.13170053337123505 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6565 |
0.13068530858255112 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
6564 |
0.13066540221414555 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6492 |
0.12923214368894467 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
6414 |
0.1276794469533104 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
6346 |
0.1263258139017318 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6294 |
0.1252906827446423 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
6176 |
0.12294173127278532 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
6169 |
0.12280238669394636 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6134 |
0.12210566379975149 |
No Hit |
| * |
6089 |
0.12120987722150094 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6084 |
0.1211103453794731 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
5972 |
0.11888083211804953 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
5948 |
0.11840307927631591 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
5902 |
0.11748738632965981 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
5761 |
0.11468058838447479 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
5737 |
0.11420283554274117 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
5703 |
0.11352601901695188 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
5630 |
0.11207285412334544 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
5393 |
0.10735504481122593 |
No Hit |
| TAACCAGAGTTAGTAGTGTATGTAAAATTCTGACTGTTCTTGCAACAATA |
5302 |
0.10554356528631928 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
5299 |
0.10548384618110256 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
5287 |
0.10524496976023576 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
5269 |
0.10488665512893554 |
No Hit |
| CATGGAGGAAGTAGGCAGCTGACAGAAAGTGCTGCAATTGCCTGTGTCAA |
5172 |
0.10295573739359548 |
No Hit |
| GACTGAACAAGTAGGAAAATGACAGAGTTATCTTCCCAATTGATGTAAGT |
5153 |
0.1025775163938897 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
5118 |
0.10188079349969484 |
No Hit |
| GCACGTGGGAGAGTAAGCAGGCCAGGTAGAGCTGCAGAAAACTAAGGCCC |
5115 |
0.10182107439447813 |
No Hit |
| AATCTCTTTCCTTTATAAATTACCCAGTCTTGCATATGTCTTTATTAGCA |
5035 |
0.10022856492203273 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality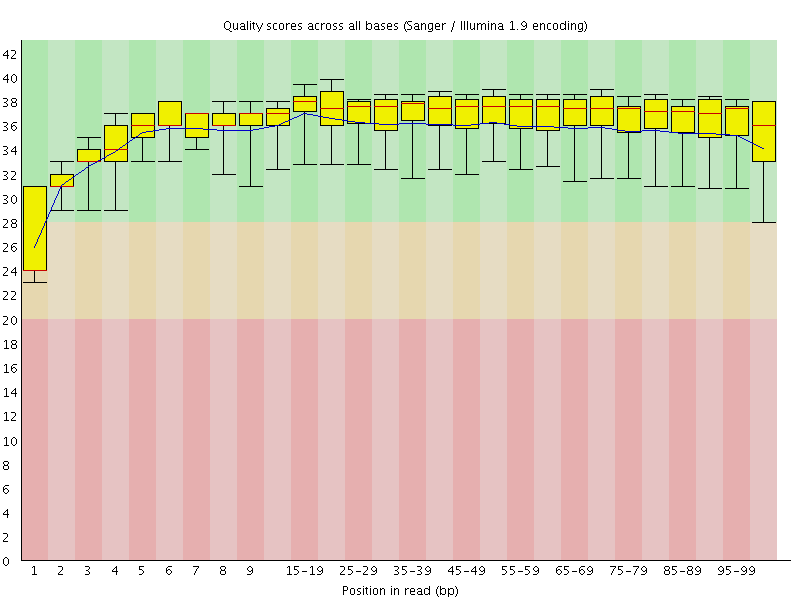
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores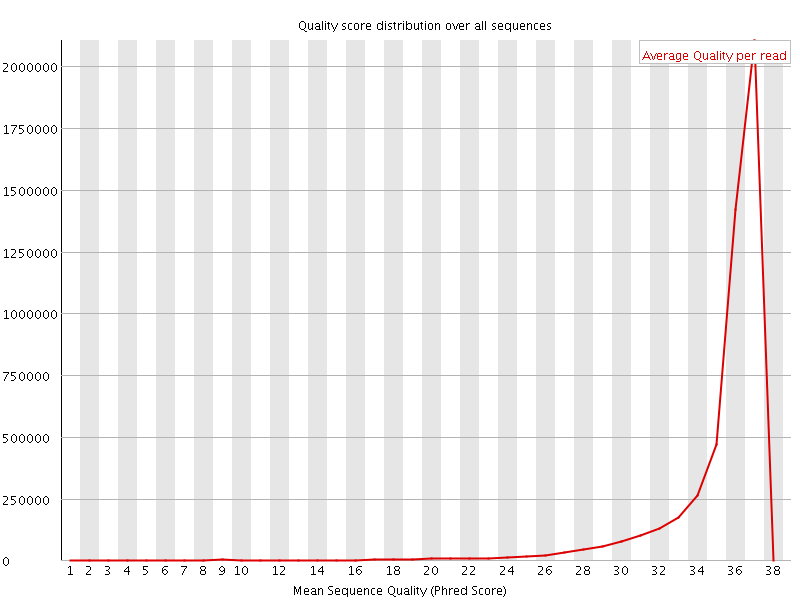
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content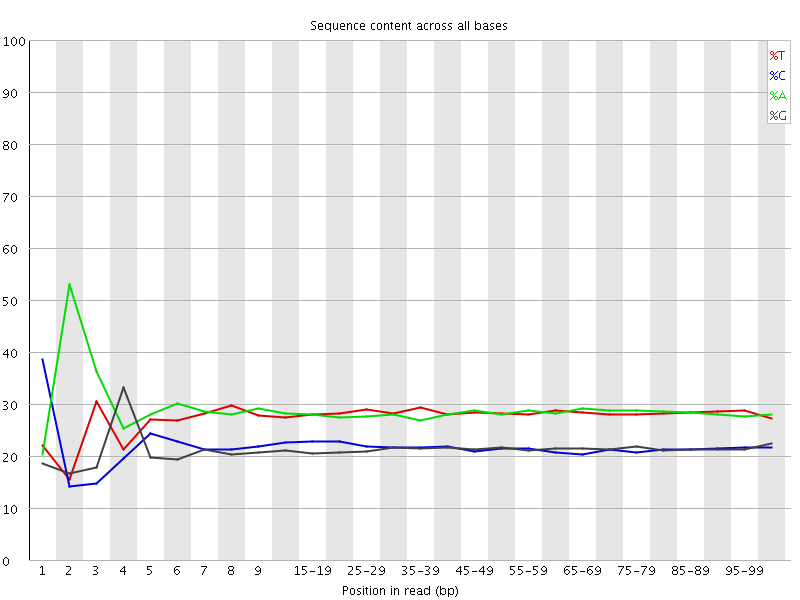
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content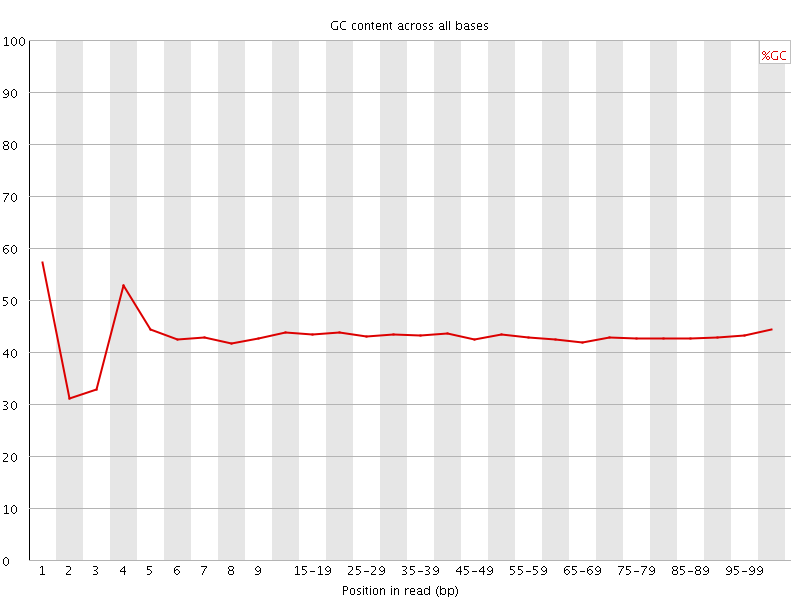
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content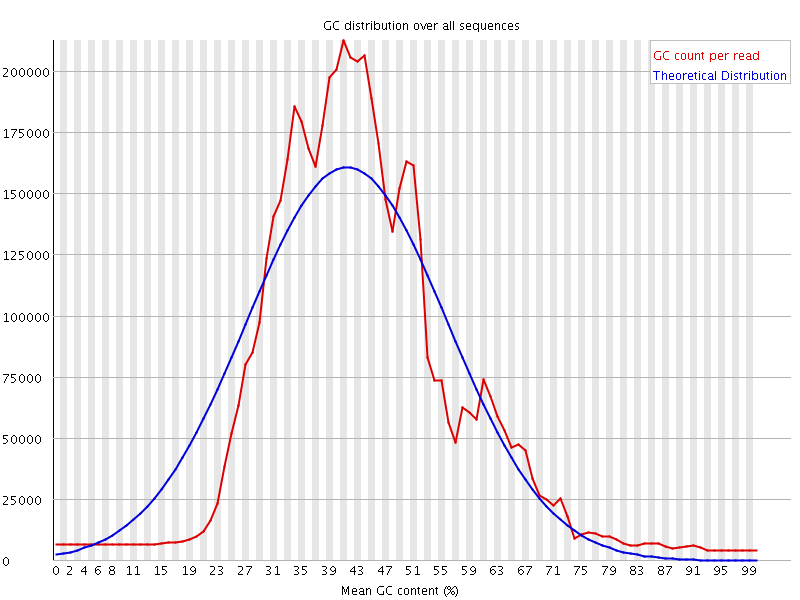
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution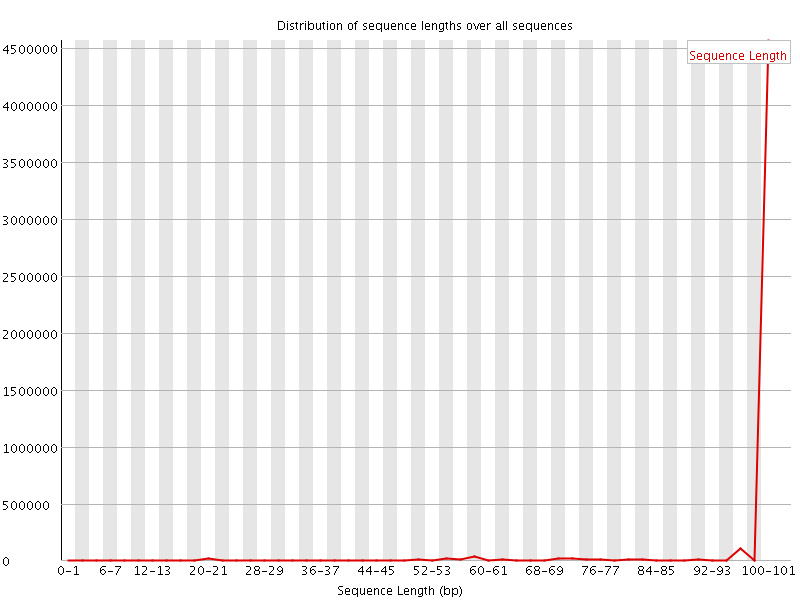
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels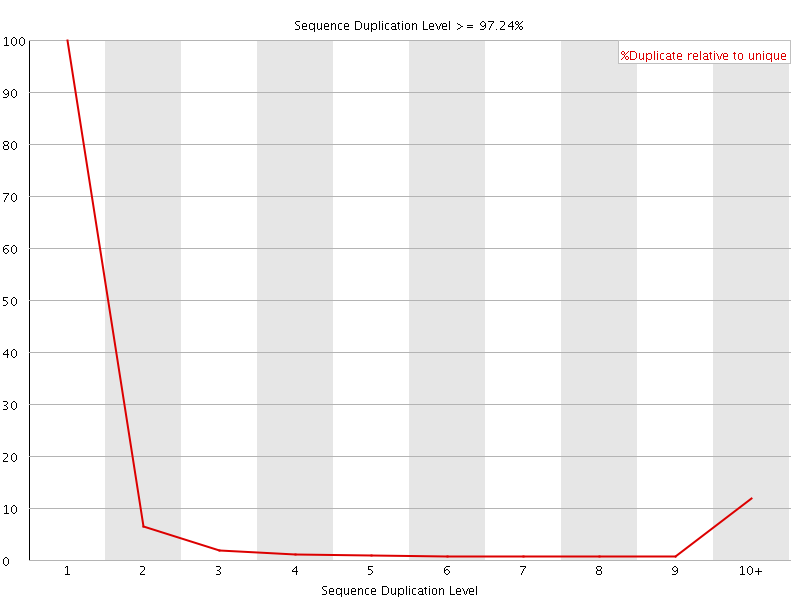
![[WARN]](Icons/warning.png) Overrepresented sequences
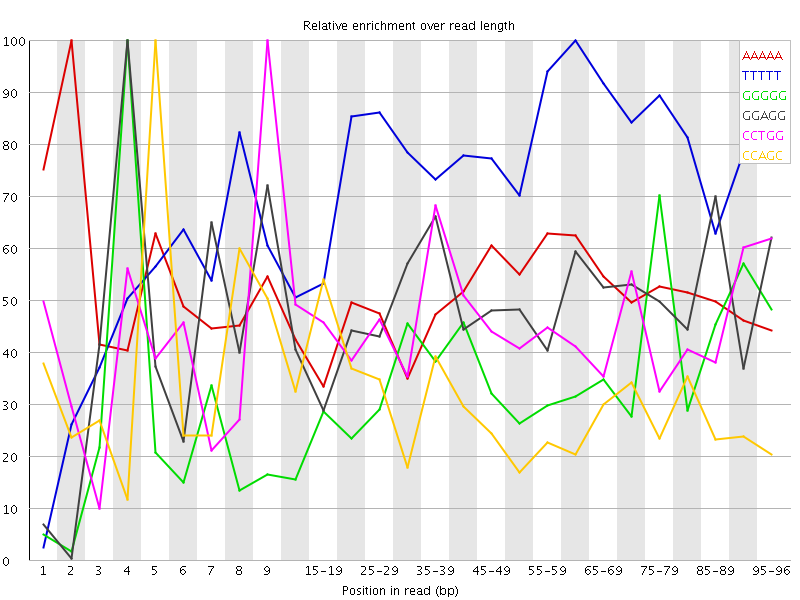
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content