| Sequence |
Count |
Percentage |
Possible Source |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
14764 |
0.23816944423473144 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
14686 |
0.23691116621723557 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
13666 |
0.22045676137305878 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
13653 |
0.2202470483701428 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
13348 |
0.21532685868634482 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
13346 |
0.2152945951474347 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
12911 |
0.20827727543447694 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
12819 |
0.20679315264461 |
No Hit |
| * |
12743 |
0.20556713816602432 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
12464 |
0.20106637448805828 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
12111 |
0.19537185987041672 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
11454 |
0.18477328733843226 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
11404 |
0.18396669886567849 |
No Hit |
| TGAGACCGCACCAGCGACACA |
10785 |
0.1739811335729869 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
10643 |
0.17169042231036621 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
10448 |
0.16854472726662653 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
10354 |
0.16702834093784946 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
10094 |
0.16283408087952989 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
9998 |
0.16128543101184264 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
9985 |
0.16107571800892667 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
9929 |
0.16017233891944246 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
9446 |
0.1523806942726411 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
9437 |
0.15223550834754543 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
8963 |
0.14458904962583974 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
8877 |
0.14320171745270327 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
8814 |
0.14218541597703352 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
8697 |
0.14029799895078973 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
8655 |
0.13962046463367656 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
8404 |
0.13557139050045267 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
8341 |
0.13455508902478291 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
8287 |
0.13368397347420885 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
8270 |
0.13340973339347256 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
8269 |
0.1333936016240175 |
No Hit |
| GCCCTCCTCCTGCCACGGGCCTGCTCCCCTCCTTCTCTCATGGGGGTCTG |
8113 |
0.13087704558902574 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
7841 |
0.12648920429724528 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
7758 |
0.125150267432474 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
7682 |
0.1239242529538883 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
7670 |
0.1237306717204274 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
7412 |
0.119568675201018 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7399 |
0.11935896219810199 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
7344 |
0.11847171487807287 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
7191 |
0.11600355415144635 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
7174 |
0.11572931407071008 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
7098 |
0.11450329959212435 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
6843 |
0.11038969838108015 |
No Hit |
| TGATCCCTTTTTCCTTTTCTCTTTCCTTGACTCCCGCTTATTCTCCTTTT |
6789 |
0.10951858283050608 |
No Hit |
| GACTGAAAGAGGGAGAAATAAAGACAAGGCCCCCGAGGAGCTGTCCAAAG |
6705 |
0.10816351419627974 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
6631 |
0.10696976325660418 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
6623 |
0.10684070910096358 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
6604 |
0.10653420548131715 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
6598 |
0.1064374148645867 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
6576 |
0.10608251593657504 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
6538 |
0.10546950869728218 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
6510 |
0.10501781915254008 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
6442 |
0.10392085882959495 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
6418 |
0.10353369636267315 |
No Hit |
| TAAACATGTCTGCAGAAGAGCAGCTGTTGCCCTTGGCCGAAAACGAGCTG |
6340 |
0.10227541834517727 |
No Hit |
| CAGGCTGGGTCCCAGCATGGCCTCAGCCTTCCTTCGCAGAAAGTTGAGCA |
6329 |
0.10209796888117145 |
No Hit |
| ATCCCCTCTTGCCATCTGTTTTTCAGAGGACCTACTAACTATGAAGAGCC |
6329 |
0.10209796888117145 |
No Hit |
| CTGAAATTATAAGTCCTCCATCACCAGACCGACCTCCTCATTCACAAACC |
6310 |
0.10179146526152502 |
No Hit |
| AGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTCTAGGCCCTGT |
6303 |
0.10167854287533948 |
No Hit |
| GTCGACAGCCAGGGGATCTCTGTGTTGGAGAATAACTGGGTGTTCGAATC |
6276 |
0.10124298510005246 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
6254 |
0.10088808617204081 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
6217 |
0.10029121070220302 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality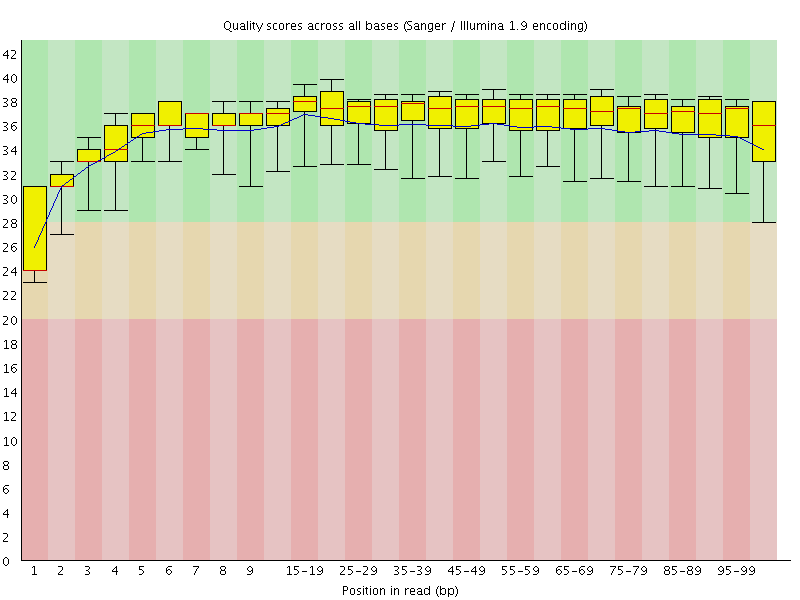
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores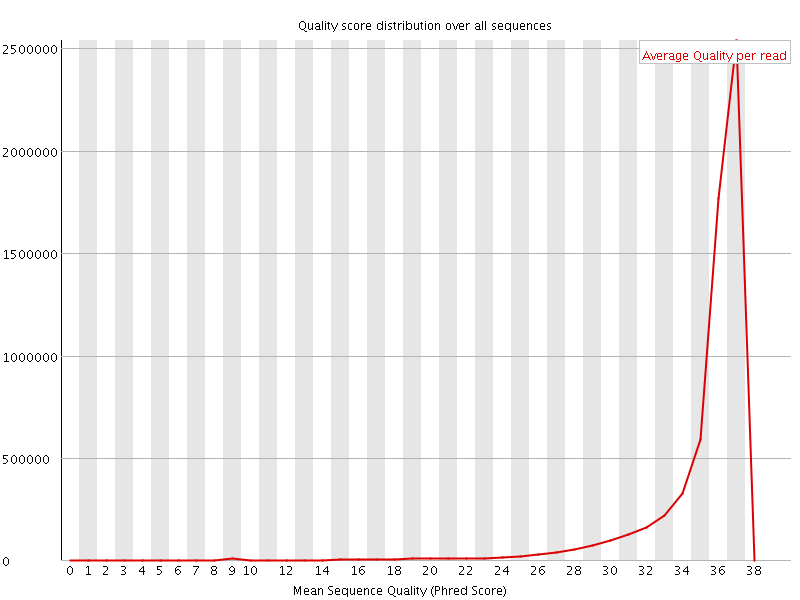
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content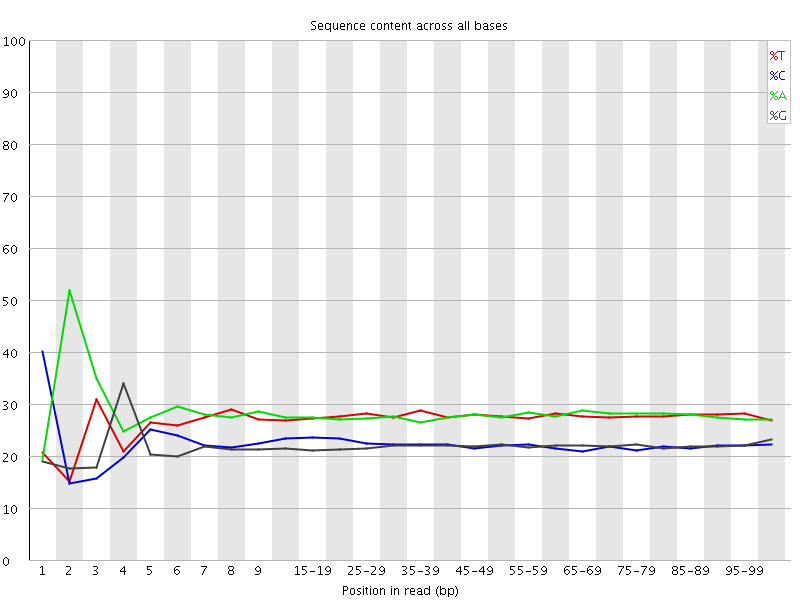
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content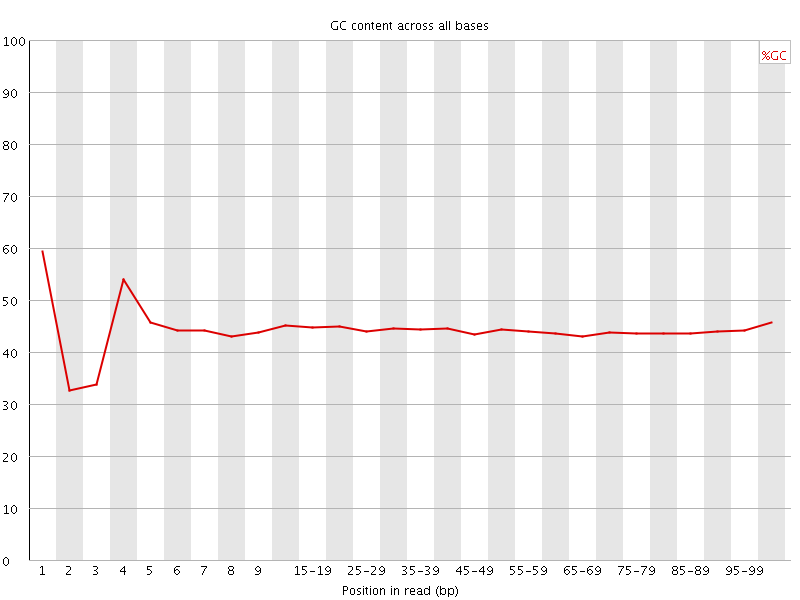
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content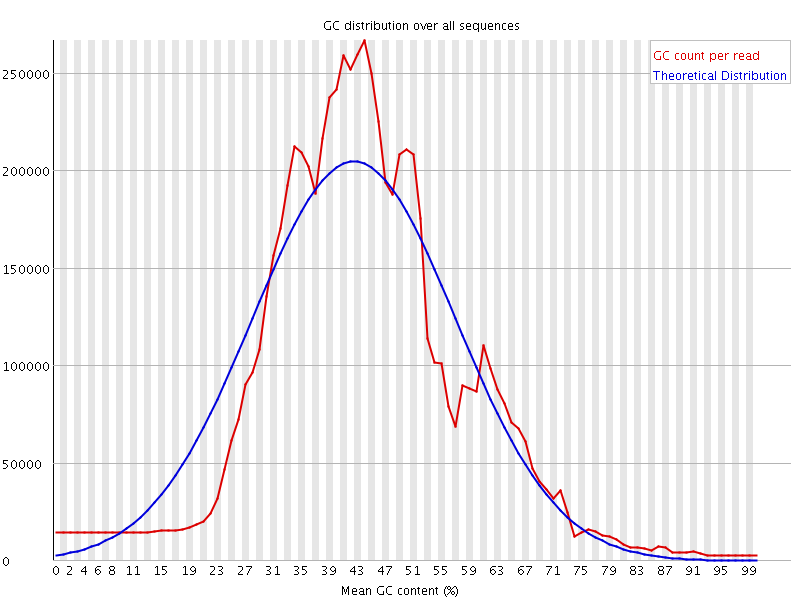
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution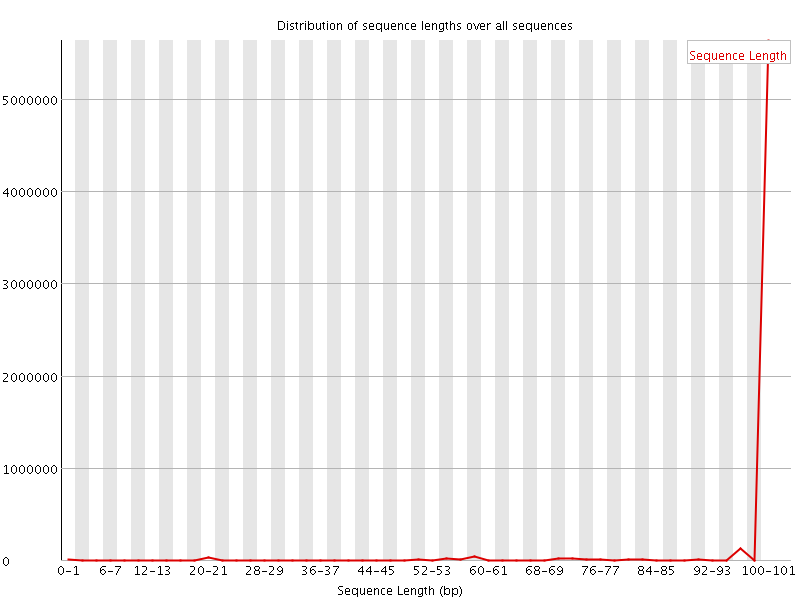
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels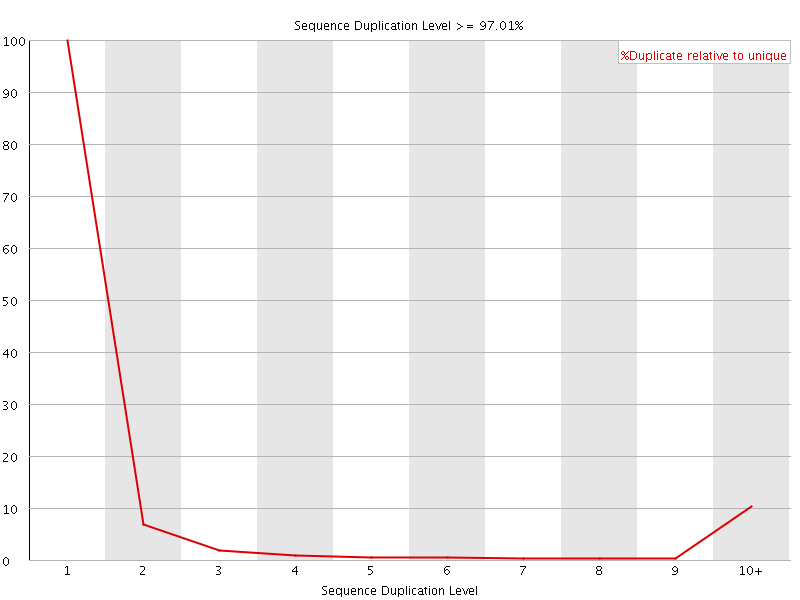
![[WARN]](Icons/warning.png) Overrepresented sequences
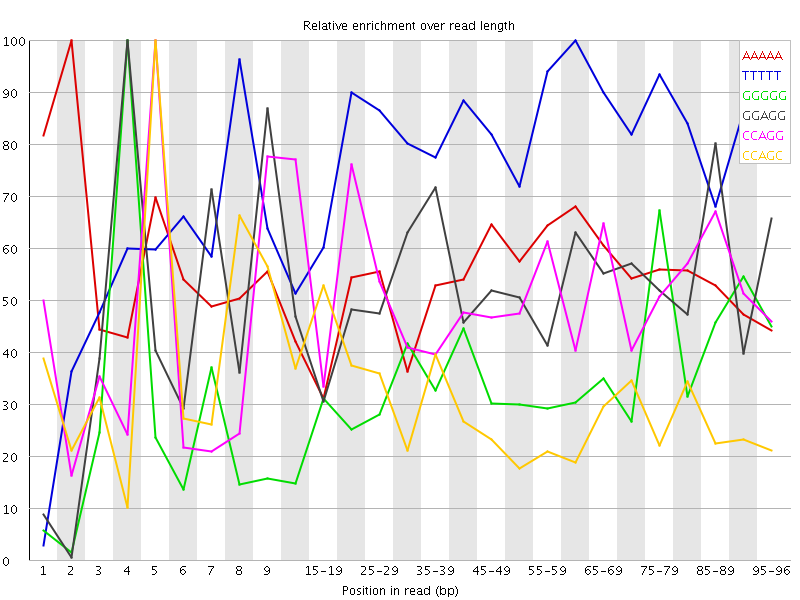
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content