| Sequence |
Count |
Percentage |
Possible Source |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
11525 |
0.2463919875249759 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
11406 |
0.2438478967210304 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
8540 |
0.1825759282831492 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
8517 |
0.18208421325381516 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
8364 |
0.17881323936302806 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8139 |
0.17400298364128236 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8025 |
0.17156578740893116 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
8010 |
0.1712451036941481 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7256 |
0.1551254022977202 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7165 |
0.15317992109470305 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
6972 |
0.14905379063116114 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
6951 |
0.14860483343046488 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
6721 |
0.14368768313712477 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
6695 |
0.14313183136483418 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
6659 |
0.14236219044935486 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
6560 |
0.14024567793178672 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
6219 |
0.1329554681490521 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
6145 |
0.1313734284894557 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
5973 |
0.1276962552266101 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
5774 |
0.12344185127715496 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
5766 |
0.123270819962604 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
5759 |
0.12312116756237192 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
5749 |
0.12290737841918321 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
5714 |
0.12215911641802277 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
5714 |
0.12215911641802277 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
5710 |
0.12207360076074729 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
5588 |
0.11946537321384516 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
5527 |
0.11816125944039409 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
5305 |
0.11341514046160496 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
5303 |
0.11337238263296723 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
5275 |
0.11277377303203888 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
5239 |
0.11200413211655955 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
5151 |
0.11012278765649901 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
5101 |
0.10905384194055551 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
5052 |
0.10800627513893088 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
4924 |
0.10526977410611552 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
4852 |
0.10373049227515689 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
4849 |
0.10366635553220027 |
No Hit |
| AGTTTGTTTGTTCATCCAGCAAATGTTTATGAGTGATGACCATGTGCCAG |
4836 |
0.10338842964605498 |
No Hit |
| GTCCAAGTACAATCCCCCAAACATTAGTAAACTACTATGTGGCGCCAAGG |
4832 |
0.10330291398877949 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
4806 |
0.10274706221648888 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
4777 |
0.10212707370124165 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
4763 |
0.10182776890077747 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
4699 |
0.1004595183843698 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
4697 |
0.10041676055573205 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality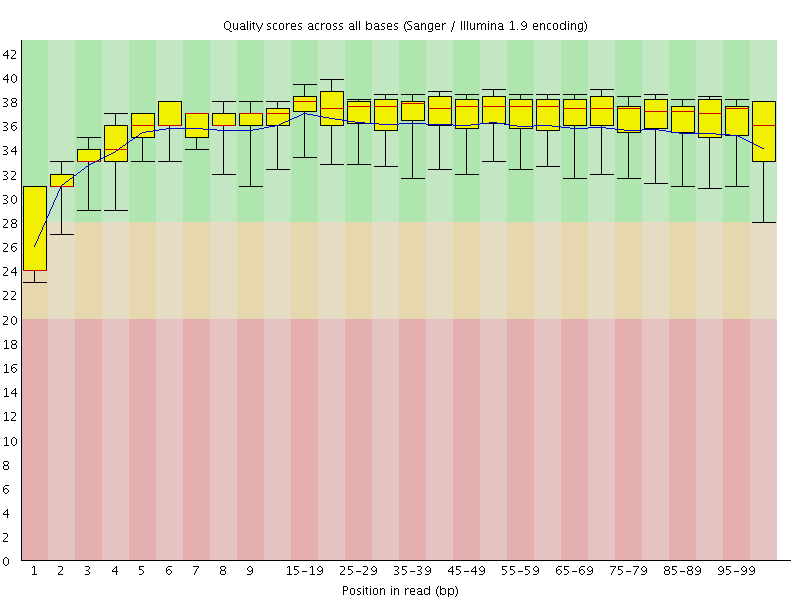
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores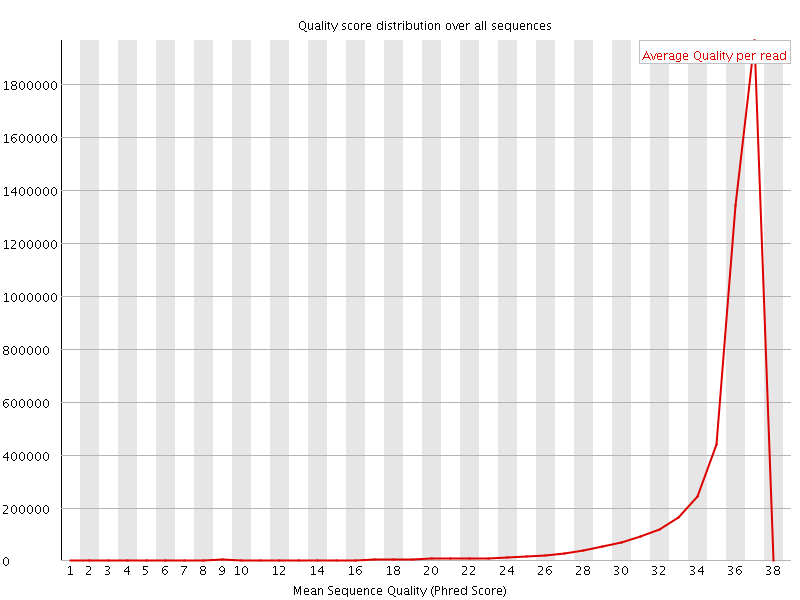
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content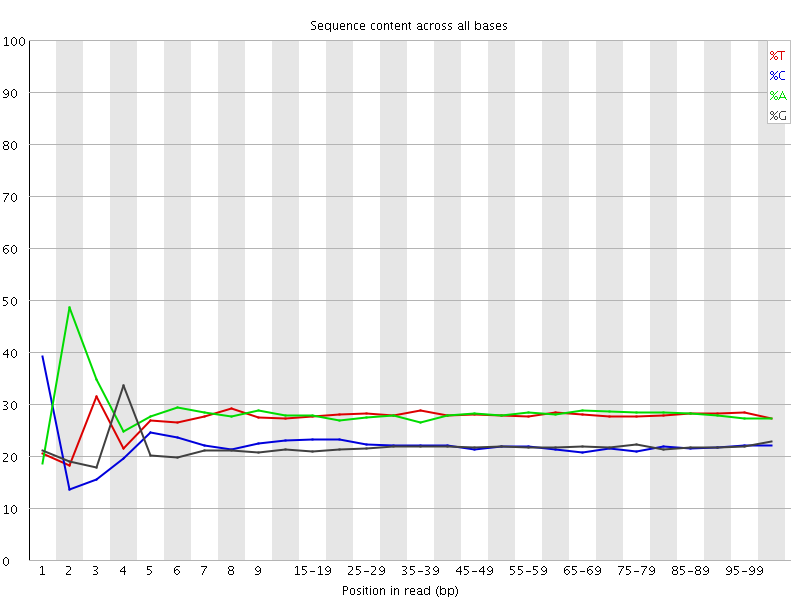
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content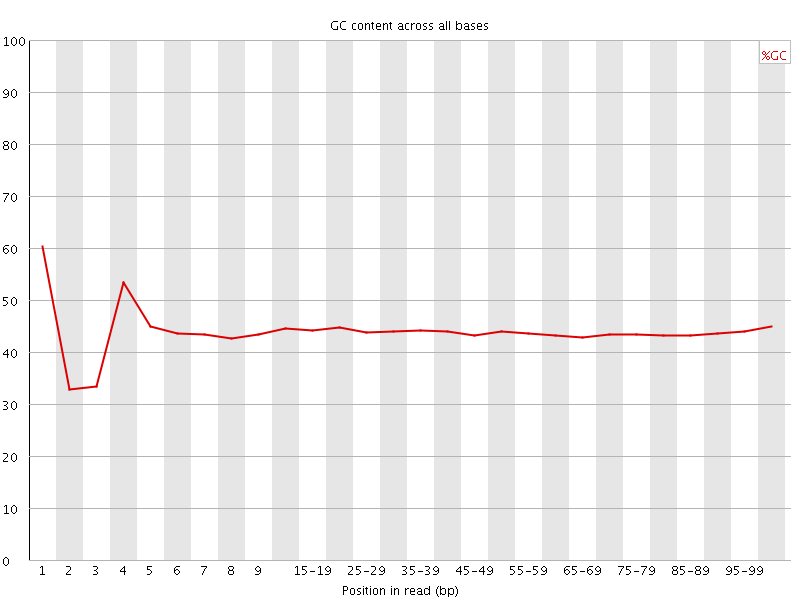
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content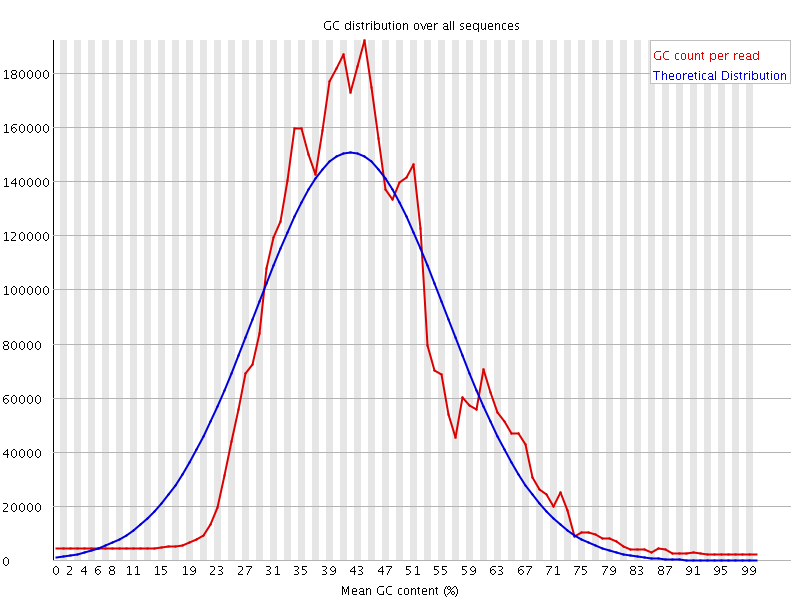
![[OK]](Icons/tick.png) Per base N content
Per base N content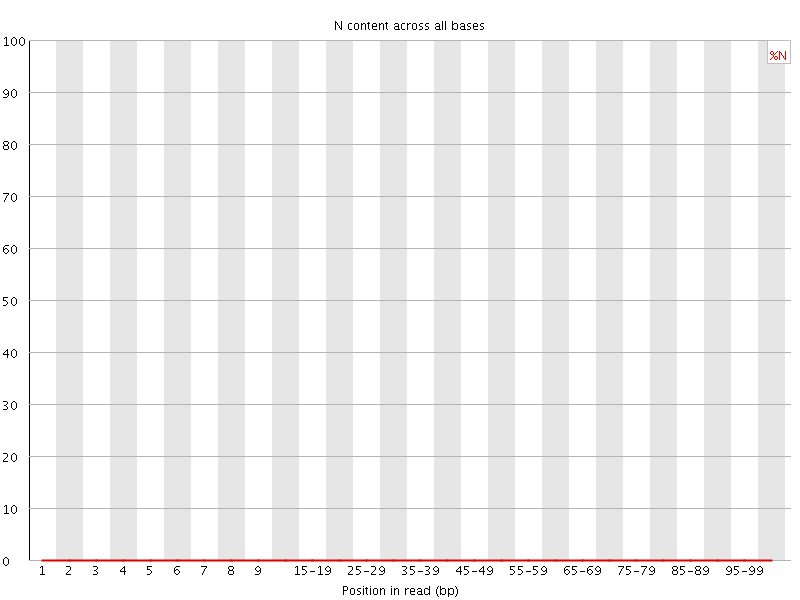
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution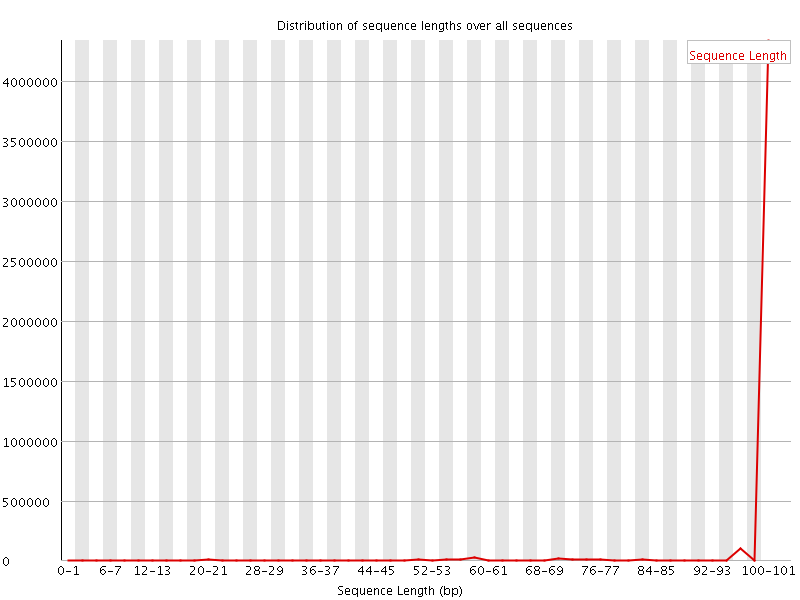
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels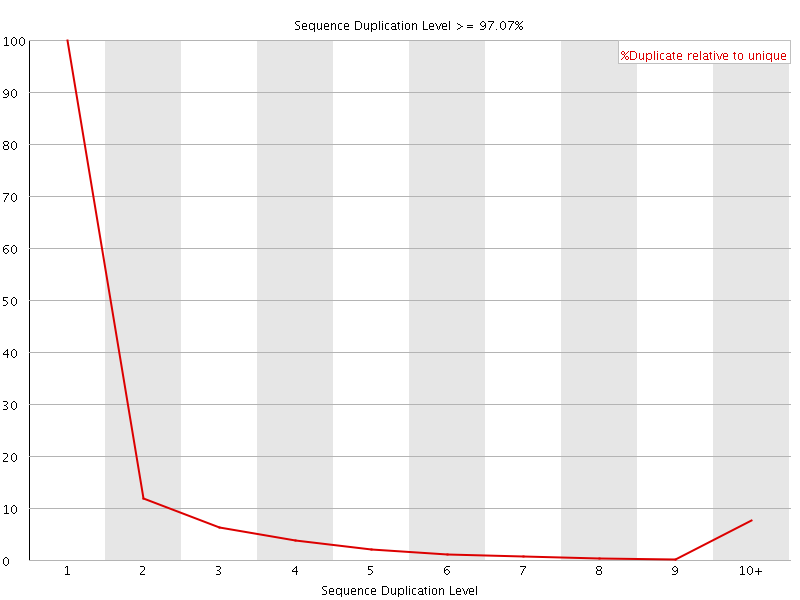
![[WARN]](Icons/warning.png) Overrepresented sequences
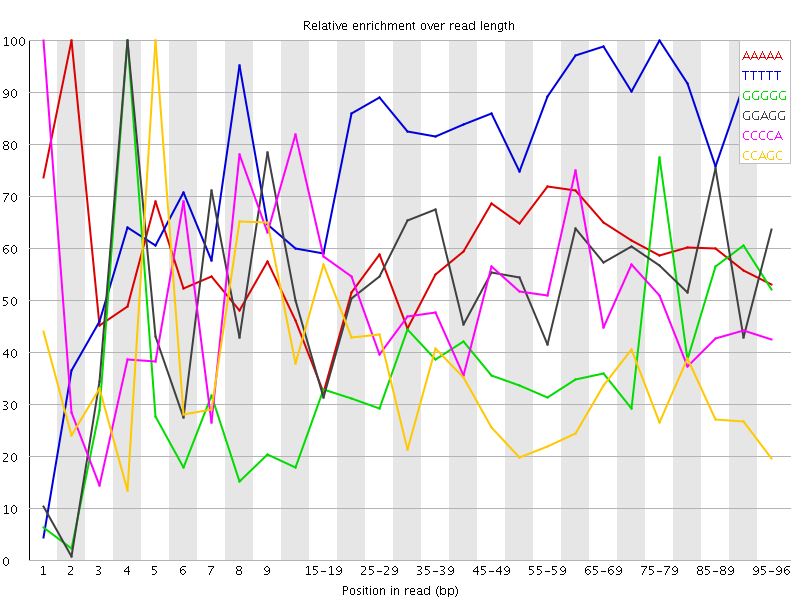
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content