| Sequence |
Count |
Percentage |
Possible Source |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
10699 |
0.2226674136781352 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
10695 |
0.22258416574330833 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
10683 |
0.22233442193882774 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
10639 |
0.22141869465573233 |
No Hit |
| TGAGACCGCACCAGCGACACA |
9402 |
0.19567427081052688 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
9287 |
0.19328089268425475 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
8763 |
0.18237541322193648 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
8762 |
0.18235460123822977 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8552 |
0.1779840846598198 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8505 |
0.17700592142560423 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
8319 |
0.1731348924561554 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
8260 |
0.17190698541745925 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
8119 |
0.16897249571481257 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
8090 |
0.16836894818731785 |
No Hit |
| TAAACATGTCTGCAGAAGAGCAGCTGTTGCCCTTGGCCGAAAACGAGCTG |
7936 |
0.16516390269648387 |
No Hit |
| ATCCCCTCTTGCCATCTGTTTTTCAGAGGACCTACTAACTATGAAGAGCC |
7886 |
0.16412330351114815 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
7859 |
0.16356137995106684 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
7840 |
0.1631659522606393 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
7800 |
0.16233347291237074 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
7763 |
0.16156342951522232 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
7717 |
0.16060607826471346 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
7689 |
0.16002334272092544 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
7656 |
0.15933654725860388 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
7647 |
0.15914923940524345 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
7637 |
0.1589411195681763 |
No Hit |
| * |
7563 |
0.15740103277387946 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
7533 |
0.15677667326267802 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
7516 |
0.1564228695396639 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7486 |
0.15579851002846246 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
7471 |
0.15548633027286177 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7389 |
0.1537797476089112 |
No Hit |
| AGAAGAGGGCTCCTTTTGTGCATATCAAACCTTGTTCCAGAATGGAGTGG |
6890 |
0.1433945677392608 |
No Hit |
| GACCCCCTCCACTAACTCCCGAGGACGTTGGCTTTGCATCTGGTTTTTCT |
6878 |
0.14314482393478023 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
6849 |
0.14254127640728553 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
6803 |
0.14158392515677667 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6738 |
0.14023114621584026 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6655 |
0.13850375156818298 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
6621 |
0.13779614412215468 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
6584 |
0.13702610072500626 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
6526 |
0.13581900567001684 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6496 |
0.1351946461588154 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
6487 |
0.135007338305455 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
6464 |
0.13452866268020058 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
6454 |
0.1343205428431334 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
6440 |
0.1340291750712394 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6433 |
0.1338834911852924 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6401 |
0.13321750770667756 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
6366 |
0.13248908827694256 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
6200 |
0.129034298981628 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
6116 |
0.12728609235026403 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6107 |
0.1270987844969036 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
6094 |
0.1268282287087163 |
No Hit |
| GATCCAAAACTTCTCTCCGACACTGAATACCTTCTGCTTCTCCTATCATT |
6061 |
0.12614143324639474 |
No Hit |
| TGATCCCTTTTTCCTTTTCTCTTTCCTTGACTCCCGCTTATTCTCCTTTT |
5994 |
0.12474703033804489 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
5986 |
0.12458053446839118 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
5982 |
0.12449728653356432 |
No Hit |
| GACTGAAAGAGGGAGAAATAAAGACAAGGCCCCCGAGGAGCTGTCCAAAG |
5955 |
0.12393536297348304 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
5756 |
0.11979377821584691 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
5743 |
0.11952322242765963 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
5592 |
0.11638061288794578 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
5560 |
0.11571462940933093 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
5556 |
0.11563138147450407 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
5535 |
0.11519432981666308 |
No Hit |
| GACCACACACCTTGGCTTATGCCTCCAACAATCCCCTTAGCATCACCATT |
5466 |
0.1137583029408998 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
5425 |
0.11290501160892451 |
No Hit |
| TGACCTAGAATGCTTTAAAGAAGTCCACCTAAATGTCGGTTCTCGCAAAA |
5406 |
0.11250958391849694 |
No Hit |
| TGATTTTTCTCTGCAGATTCCCCAGACTGAGTCAGGGAATGAGAAGGAAA |
5393 |
0.11223902813030966 |
No Hit |
| GCCCTCCTCCTGCCACGGGCCTGCTCCCCTCCTTCTCTCATGGGGGTCTG |
5353 |
0.11140654878204108 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
5216 |
0.10855530701422125 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
5192 |
0.1080558194052601 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
5186 |
0.10793094750301982 |
No Hit |
| CAGAGAAAAATCAGAGACCAAGGAAGCAGACTAGTGCTCCGGCAGAGCCA |
5185 |
0.10791013551931311 |
No Hit |
| CCTTGTCTTTATTTCTCCCTCTTTCAGTCTGAGAGCCTGGGGTAAACCAA |
5185 |
0.10791013551931311 |
No Hit |
| TGAGGAGCGGAGCGATACCCCTGAAGTTCATCCTCCACTGCCCATTTCCC |
5015 |
0.1043720982891717 |
No Hit |
| TAACCAGAGTTAGTAGTGTATGTAAAATTCTGACTGTTCTTGCAACAATA |
5002 |
0.10410154250098441 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
4982 |
0.10368530282685012 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
4973 |
0.1034979949734897 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
4912 |
0.10222846396738014 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
4911 |
0.10220765198367342 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
4888 |
0.10172897635841897 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
4853 |
0.101000556928684 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
4825 |
0.100417821384896 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
4805 |
0.10000158171076172 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality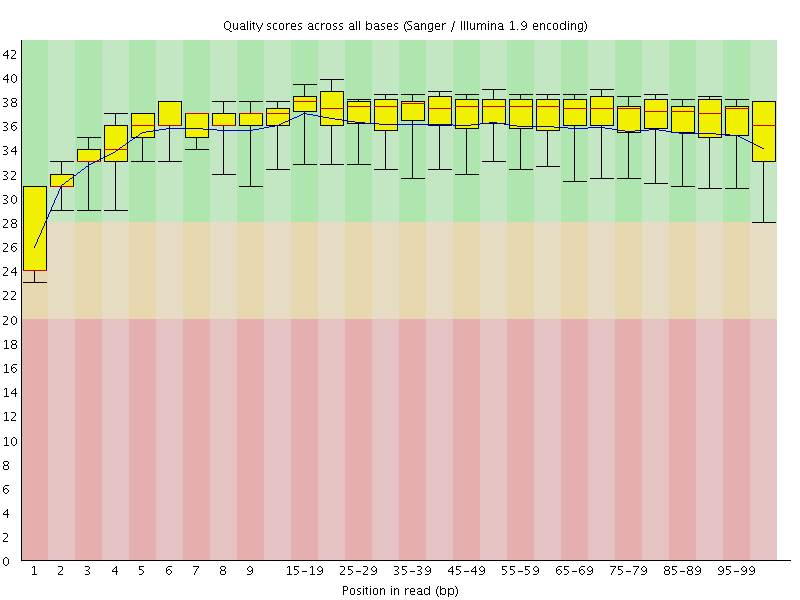
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores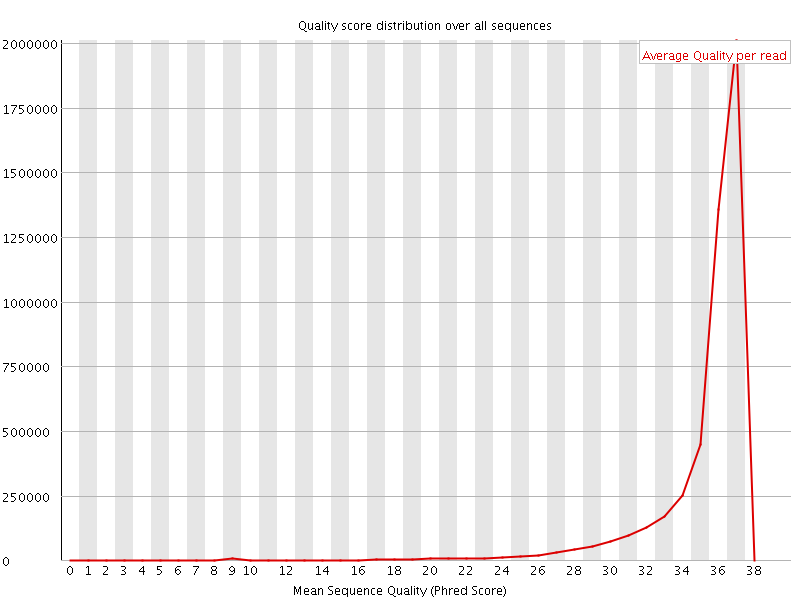
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content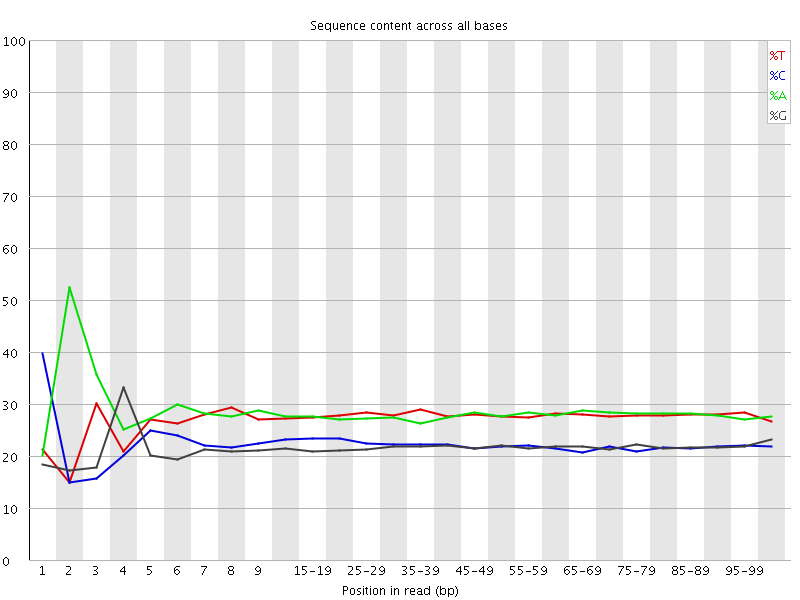
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content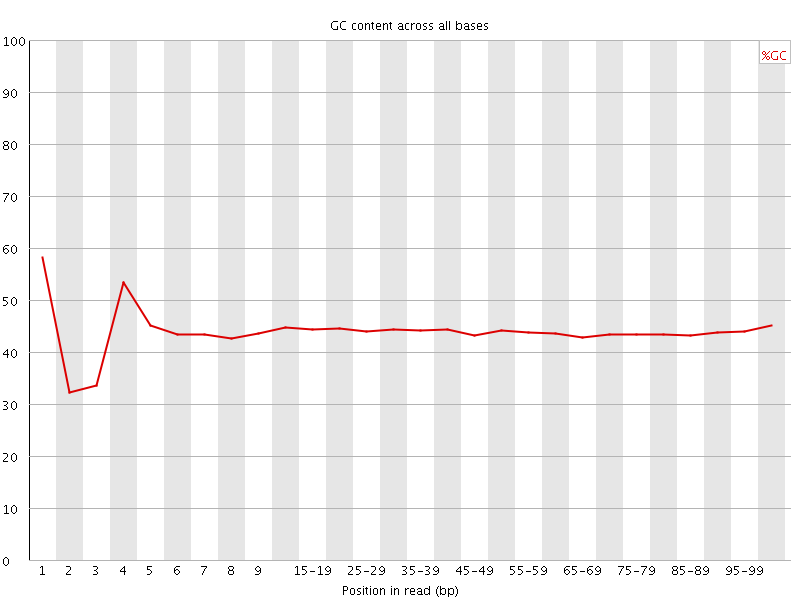
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content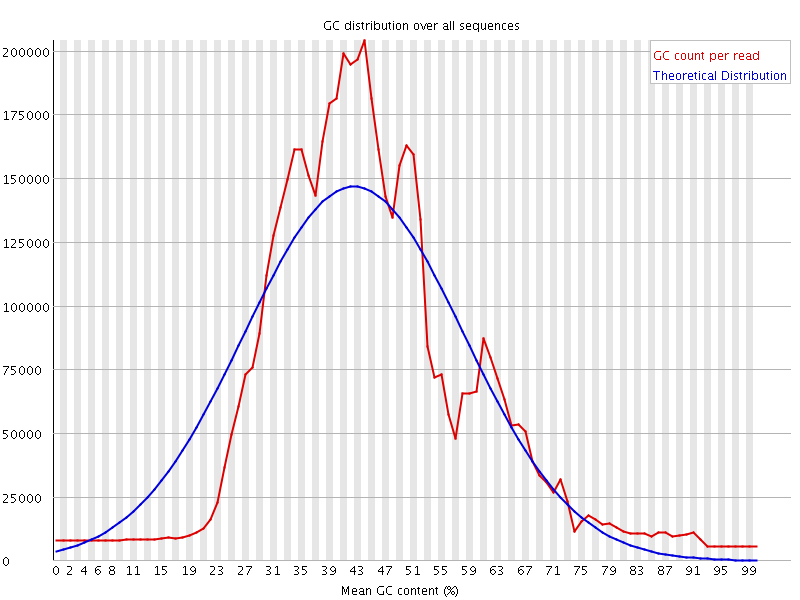
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution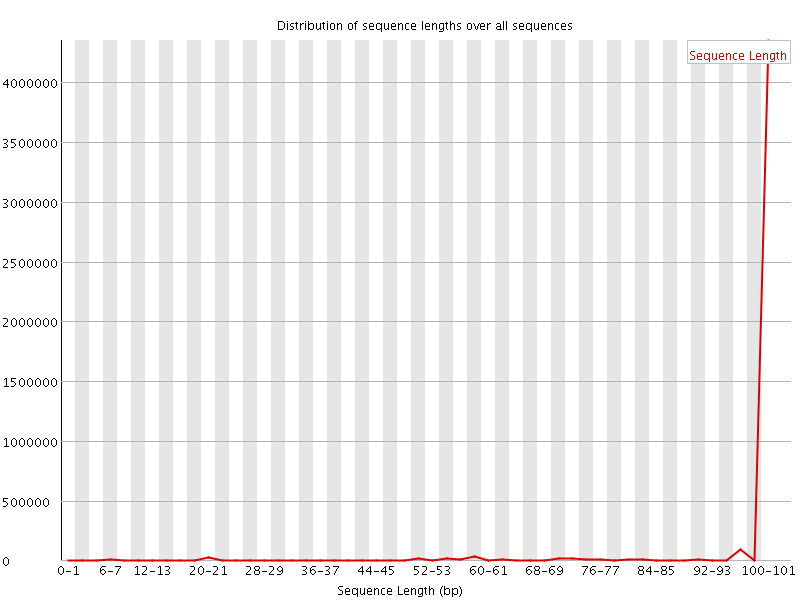
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels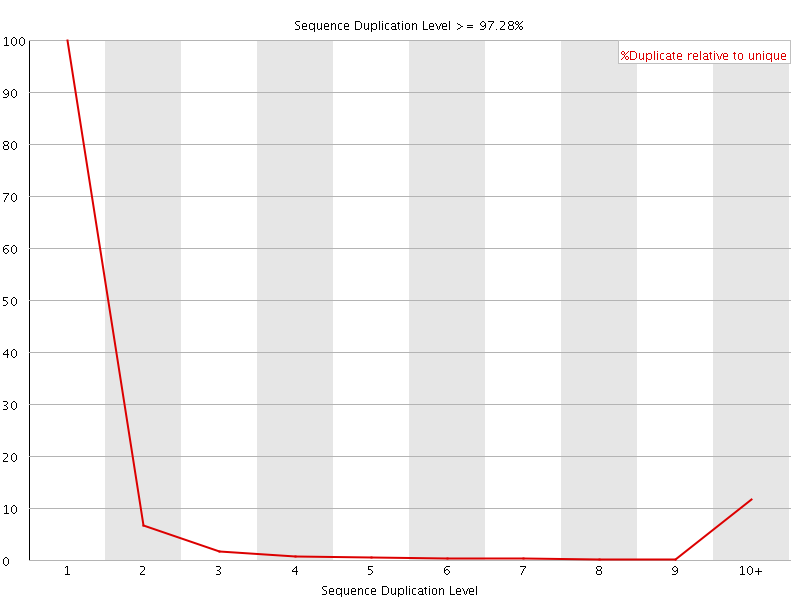
![[WARN]](Icons/warning.png) Overrepresented sequences
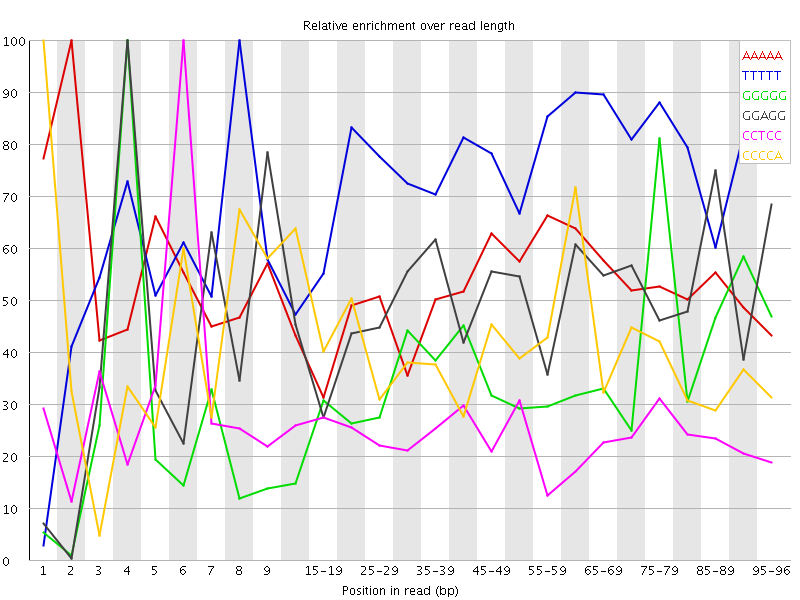
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content