| Sequence |
Count |
Percentage |
Possible Source |
| * |
15058 |
0.29891182438642483 |
No Hit |
| TGAGACCGCACCAGCGACACA |
12954 |
0.25714595385188915 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
12816 |
0.2544065574004795 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
11795 |
0.23413899379983272 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
11746 |
0.23316630955259304 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
11700 |
0.23225317740212312 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
11637 |
0.23100258336995788 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
11458 |
0.22744930826269463 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
11405 |
0.2263972212197619 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
9090 |
0.18044285321241876 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
8911 |
0.1768895781051555 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
8797 |
0.17462659842790407 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
8758 |
0.17385242116989696 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
8669 |
0.17208570896572697 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
8622 |
0.17115272611633384 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8545 |
0.16962422229924293 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
8529 |
0.1693066111164708 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8424 |
0.16722228772952866 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
8371 |
0.16617020068659596 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
8369 |
0.16613049928874946 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
8347 |
0.16569378391243778 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
8282 |
0.16440348848242597 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
8276 |
0.16428438428888642 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
8200 |
0.16277573117071878 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
8156 |
0.16190230041809542 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
7455 |
0.1479869604728913 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
7429 |
0.14747084230088658 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
7225 |
0.14342129972054185 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
7202 |
0.14296473364530693 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6913 |
0.13722788165648525 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6776 |
0.13450833590399885 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
6736 |
0.1337143079470685 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
6709 |
0.13317833907614052 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6684 |
0.1326820716030591 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6682 |
0.13264237020521255 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6651 |
0.13202699853859154 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6432 |
0.12767969547439795 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
6339 |
0.12583358047453494 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6337 |
0.1257938790766884 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
6301 |
0.1250792539154511 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6141 |
0.12190314208772976 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
6098 |
0.12104956203402965 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
6042 |
0.11993792289432718 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
5991 |
0.11892553724924101 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
5975 |
0.11860792606646886 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
5961 |
0.11833001628154326 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
5943 |
0.11797270370092461 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
5898 |
0.11707942224937799 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
5716 |
0.11346659504534494 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
5698 |
0.1131092824647263 |
No Hit |
| GTATCTTAATGATGTATATAATTGCCTTCAATCCCCTTCTCACCCCACCC |
5648 |
0.11211674751856339 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
5648 |
0.11211674751856339 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
5620 |
0.11156092794871214 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
5601 |
0.11118376466917024 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
5573 |
0.110627945099319 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
5568 |
0.11052869160470272 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
5544 |
0.1100522748305445 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
5537 |
0.10991331993808169 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
5494 |
0.10905973988438158 |
No Hit |
| CATGACTATAAATATCAAGCAAATATGATTTTACTCACAAGTTCTGGAAT |
5407 |
0.1073327290780581 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
5390 |
0.10699526719636272 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
5365 |
0.10649899972328125 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATCTGGGTTTTTTTC |
5354 |
0.10628064203512541 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
5343 |
0.10606228434696957 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
5331 |
0.10582407595989046 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
5294 |
0.10508960009972992 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
5271 |
0.10463303402449496 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
5222 |
0.10366034977725531 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
5203 |
0.10328318649771341 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
5196 |
0.10314423160525057 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
5193 |
0.10308467950848081 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
5186 |
0.102945724616018 |
No Hit |
| AGTTCTAACATTTTGTTTATGGCTTTTTGGGGGGTTTTCATTGCAGGATG |
5145 |
0.1021318459601644 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
5145 |
0.1021318459601644 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
5122 |
0.10167527988492947 |
No Hit |
| TAAGCAATGCCACAGGGGCATGATATCCTGGCTTCTTCTGACCAAGCAAA |
5121 |
0.10165542918600622 |
No Hit |
| AGAACAGTTGCAAATAGACTACATAATATACGTGGGCAAAAAGGCAATTA |
5112 |
0.10147677289569687 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
5091 |
0.10105990821830846 |
No Hit |
| TAATTGCCTTTTTGCCCACGTATATTATGTAGTCTATTTGCAACTGTTCT |
5073 |
0.1007025956376898 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality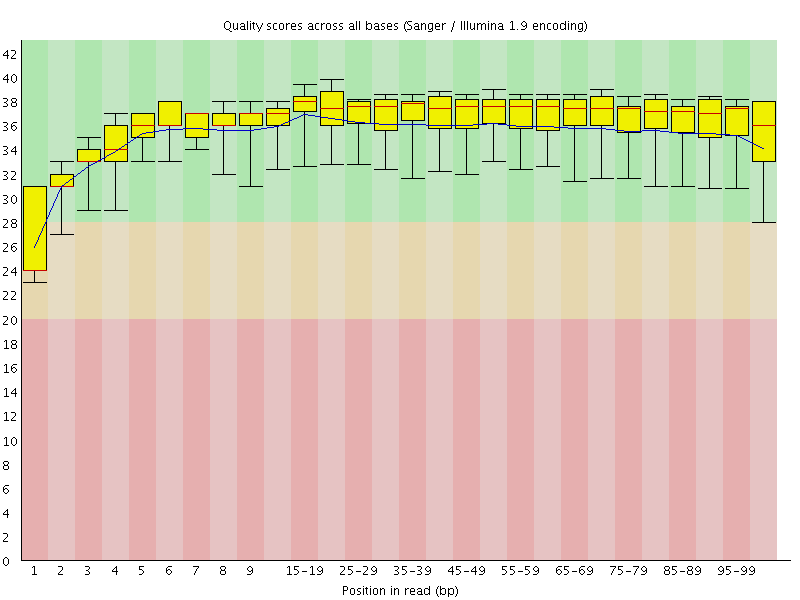
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores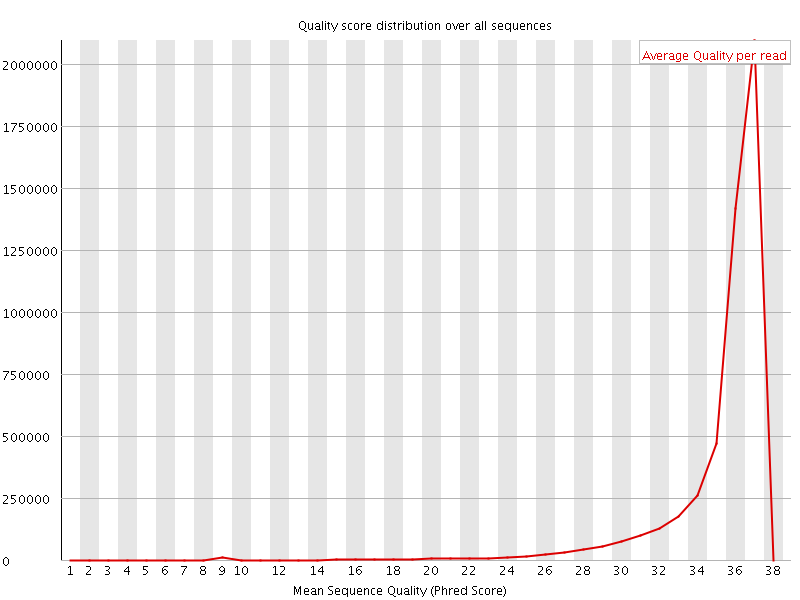
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content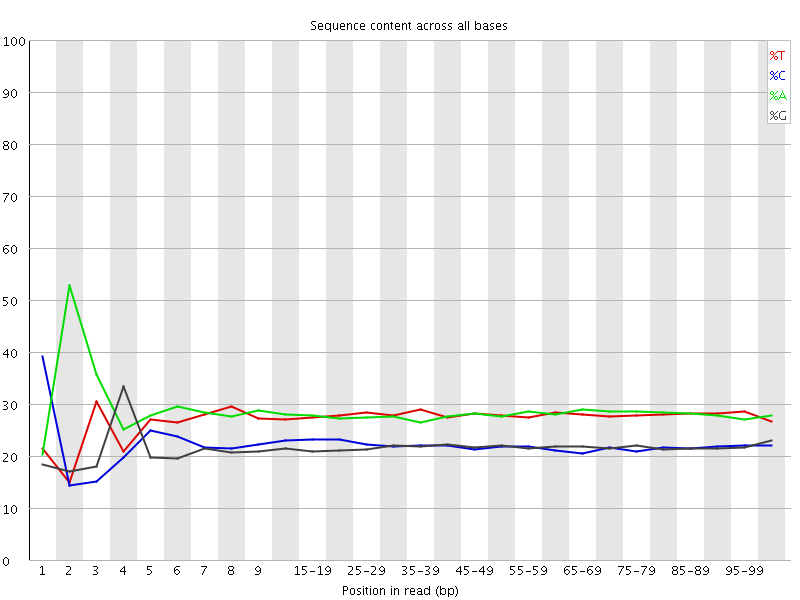
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content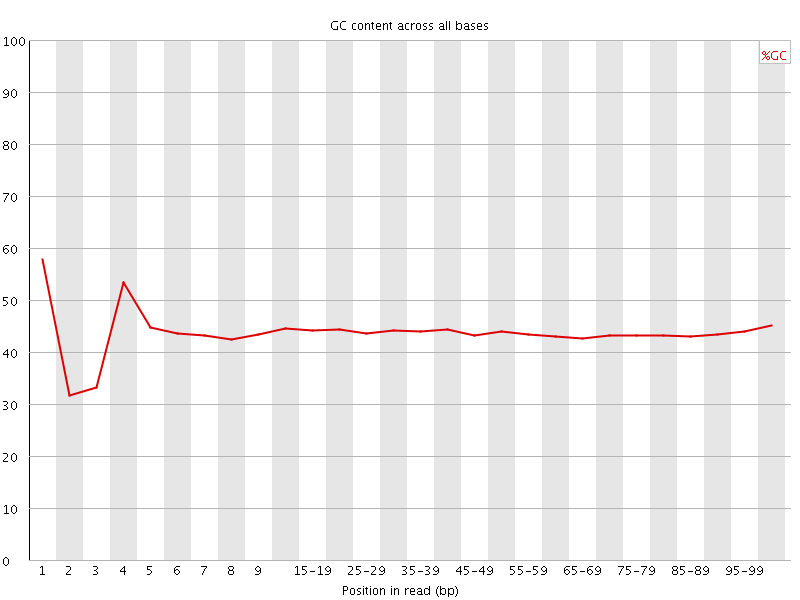
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content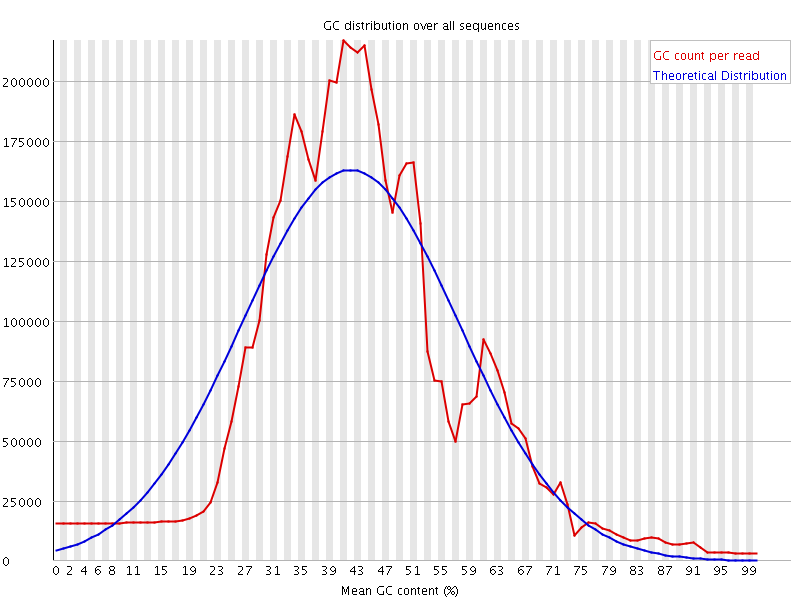
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution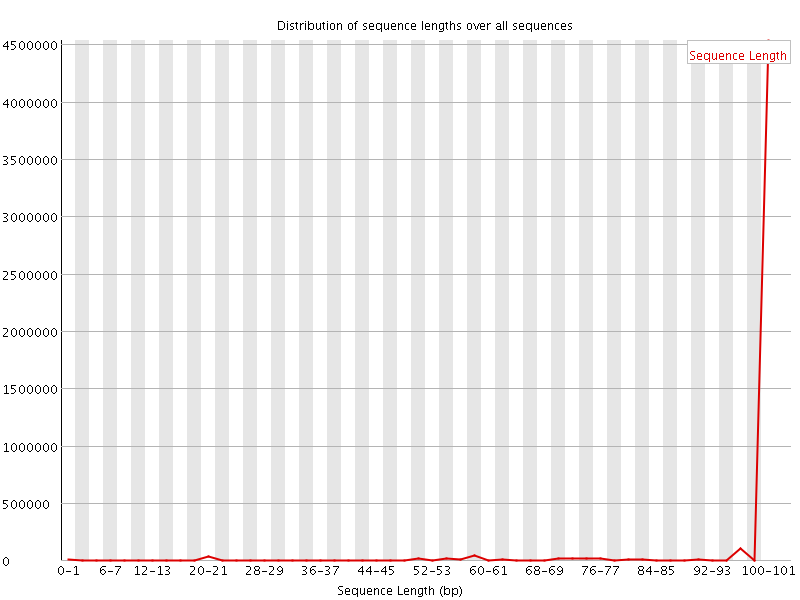
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels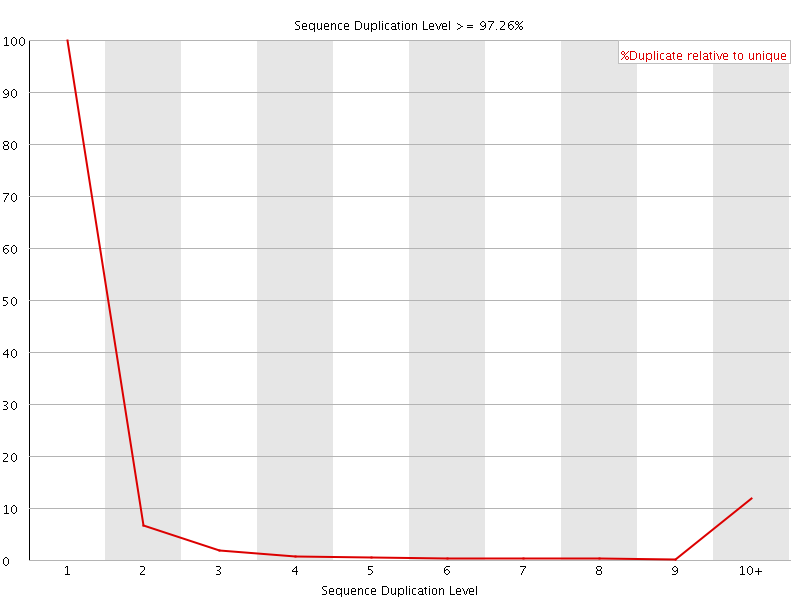
![[WARN]](Icons/warning.png) Overrepresented sequences
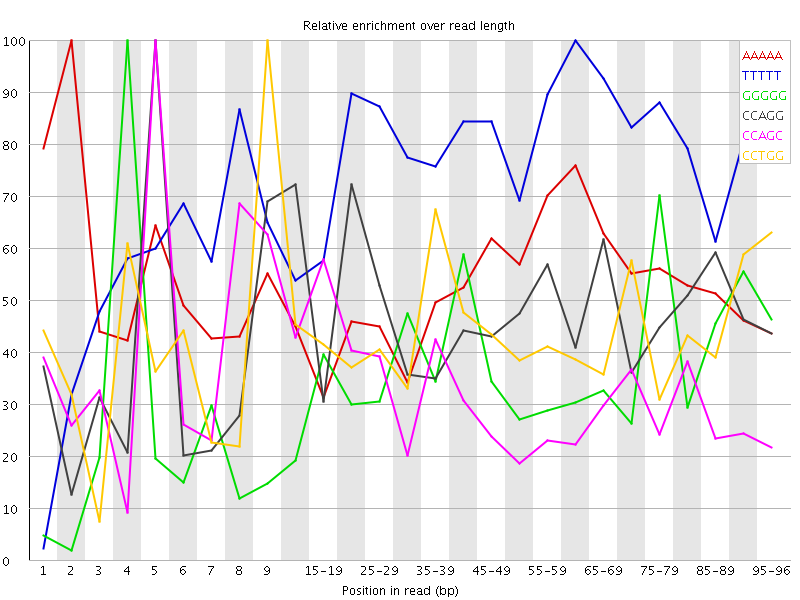
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content