| Sequence |
Count |
Percentage |
Possible Source |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
30776 |
0.4965749943446343 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
30616 |
0.4939933723308852 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
16466 |
0.2656811754899515 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
15731 |
0.2538218493642917 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
15720 |
0.25364436285084646 |
No Hit |
| CAGGCTGGGTCCCAGCATGGCCTCAGCCTTCCTTCGCAGAAAGTTGAGCA |
13999 |
0.22587579106545802 |
No Hit |
| AGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTCTAGGCCCTGT |
13953 |
0.22513357473650514 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
13930 |
0.22476246657202872 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
13626 |
0.2198573847459055 |
No Hit |
| CGAGACCGCGCCCCAGGCCCAGCACTCACAAATTCCTGGTCGTGGTTATT |
13105 |
0.21145097806363503 |
No Hit |
| GGCACAAGGGTACCTACGGGCTGCTGCGGCGGCGAGAGGACTGGCCCTCC |
12985 |
0.20951476155332324 |
No Hit |
| ATACTTGTAATTCCCCTAACCATAAGATTTACTGCTGCTGTGGATATCTC |
12471 |
0.20122130083415432 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
11649 |
0.18795821773851848 |
No Hit |
| CTGAGGAAATAAGATGTAGGGCATTTCTGATGTGACTCAGTGGGAAAACT |
11644 |
0.18787754205058882 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
11635 |
0.18773232581231544 |
No Hit |
| TGCTCTGTGGTCCGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTG |
11544 |
0.18626402829199565 |
No Hit |
| CCGCGGCGCAGATCGAAACAGATCGGGCGGCTCGGGTTACACACGCACGC |
11468 |
0.18503775783546483 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
11271 |
0.1818591357310363 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
11192 |
0.18058445986174768 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
9425 |
0.15207367174740635 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
9382 |
0.1513798608312113 |
No Hit |
| AATCTCTTTCCTTTATAAATTACCCAGTCTTGCATATGTCTTTATTAGCA |
8906 |
0.1436995353403078 |
No Hit |
| CGGACGGGTCGGTGGCGTAGACGCGGAGCGCGCAGCTCACACCTGGCGGC |
8846 |
0.1427314270851519 |
No Hit |
| GGCTCCGCGGAGGCGCCACGCACAGTGTCCAAAGCATCCTTGCTTCCTCC |
8805 |
0.14206988644412868 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
8715 |
0.14061772406139483 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
8681 |
0.14006912938347316 |
No Hit |
| CATGCCCACAGAGACTTGCACAACATGCAGAATGGCAGCACATTGGTAAG |
8669 |
0.13987550773244198 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
8660 |
0.1397302914941686 |
No Hit |
| ATAGGACCTTCAGTCGGCAACGTGTTCTCAGTGGGTTCCAGCTTGTGTGG |
8548 |
0.13792315608454425 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
8409 |
0.13568037196009974 |
No Hit |
| ACTACTGTACAACCCGACTTCATAATGGTGCTTTCAAACAGCGAGATGAG |
8325 |
0.13432502040288147 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
8285 |
0.13367961489944422 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
8272 |
0.1334698581108271 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
8219 |
0.13261469581877272 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
8149 |
0.1314852361877575 |
No Hit |
| AAGAAATGTGTACTGCTAAATGTGAACTGCTACTTTTTTCTAAGTAGTTT |
7957 |
0.1283872897712586 |
No Hit |
| CACCGTGAGAACGCGCCATCTGCAAGCTGTCTCCCTTTCTCCATCCTTGG |
7951 |
0.12829047894574303 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
7940 |
0.12811299243229776 |
No Hit |
| CTGGGGTCTCAACGCACACCCGAGGTCTTCCGGGGATTCTGATACCTACG |
7928 |
0.1279193707812666 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7913 |
0.1276773437174776 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
7900 |
0.12746758692886048 |
No Hit |
| CTGGAGGACCGAGGTATACAAGAAGTGTTGCCATTTATCAAAAGGTCATC |
7784 |
0.12559591096889242 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTATCTGCATTTCCC |
7430 |
0.1198840722634726 |
No Hit |
| AGAACAGTTGCAAATAGACTACATAATATACGTGGGCAAAAAGGCAATTA |
7425 |
0.11980339657554294 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAAAAGTAATTCTGGCCTGGCAACCTA |
7376 |
0.11901277483383227 |
No Hit |
| TAATTGCCTTTTTGCCCACGTATATTATGTAGTCTATTTGCAACTGTTCT |
7364 |
0.1188191531828011 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
7244 |
0.1168829366724893 |
No Hit |
| TAAAAATTTACAGATTGTGCTGAGCTTGACAAATAAGTGTATCCTTATGT |
7179 |
0.11583415272940374 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
7145 |
0.11528555805148206 |
No Hit |
| TGCTCTAAAATTTCATTTATCATACTGAAATATACTCACCAGAATATGGG |
7107 |
0.11467242282321664 |
No Hit |
| TAACCAAAGTCTTGCCACAAGACAAAGAATACTATAAAGTAAAAGAACCT |
7028 |
0.11339774695392806 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
7007 |
0.11305890906462349 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
6983 |
0.11267166576256113 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
6959 |
0.11228442246049876 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
6946 |
0.11207466567188164 |
No Hit |
| ACCCTATATATTCCCTTTATTATCTATGAAAACGTCTAGATGATTTCCTA |
6945 |
0.11205853053429572 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
6933 |
0.11186490888326454 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
6909 |
0.11147766558120217 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
6901 |
0.11134858448051473 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
6896 |
0.11126790879258508 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
6875 |
0.11092907090328051 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
6866 |
0.11078385466500712 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
6859 |
0.1106709087019056 |
No Hit |
| TAATTATGGAATATTTACCATATGGAAGTTTACGAGACTATCTTCAAAAA |
6857 |
0.11063863842673373 |
No Hit |
| ATCCCCTCTTGCCATCTGTTTTTCAGAGGACCTACTAACTATGAAGAGCC |
6852 |
0.11055796273880408 |
No Hit |
| TAAACATGTCTGCAGAAGAGCAGCTGTTGCCCTTGGCCGAAAACGAGCTG |
6818 |
0.11000936806088239 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
6815 |
0.10996096264812459 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
6812 |
0.1099125572353668 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
6772 |
0.10926715173192954 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
6700 |
0.10810542182574244 |
No Hit |
| GGCACCCTTTTCTCGCCGCAAGAAGCCCATTCCTATGGAAGTCTAGCAAA |
6645 |
0.1072179892585162 |
No Hit |
| CAGGATGGAAGGCACAGCAGAGCCATGAGTCCTGGGGCAGAGAGTGCTGG |
6599 |
0.10647577292956334 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
6580 |
0.10616920531543064 |
No Hit |
| AAGAAGTCCACCTAAATGTCGGTTCTCGCAAAATAGATTTTCGCTTCCCT |
6523 |
0.10524950247303252 |
No Hit |
| ATCATGTCCCCAGAAAATCCAGTTACCTCTTGGTCAGTCATCCGGAAGCA |
6499 |
0.10486225917097018 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
6491 |
0.1047331780702827 |
No Hit |
| TAATAGGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGT |
6444 |
0.10397482660374394 |
No Hit |
| TAACCAGAGTTAGTAGTGTATGTAAAATTCTGACTGTTCTTGCAACAATA |
6423 |
0.10363598871443937 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
6404 |
0.10332942110030667 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
6381 |
0.10295831293583024 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
6374 |
0.10284536697272872 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
6329 |
0.10211928578136178 |
No Hit |
| ACTGAGTCACACTGCATAGGAATTTAGAACCTAACTTTTATAGGTTATCA |
6314 |
0.1018772587175728 |
No Hit |
| TAACATGCCCCACAAAGTTTCTATGTATATATTAGGACAAAATTGTGCAA |
6300 |
0.10165136679136975 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
6293 |
0.10153842082826824 |
No Hit |
| AGGACAACAGCCGATCCATAAATTTGCTGACAGGTGTATCTGCGTTTCCC |
6276 |
0.10126412348930741 |
No Hit |
| CCCCAGCAGCAGACCTCCTCGTCTCCACCTCCACCTCTGCTGACTCCACC |
6275 |
0.10124798835172148 |
No Hit |
| CTCCTCCTCCAGCTCACTGCTGCTGCTGCCGCTGCCACCGCTTCTGCTGC |
6262 |
0.10103823156310436 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
6260 |
0.1010059612879325 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
6247 |
0.10079620449931538 |
No Hit |
| AGTTCTAACATTTTGTTTATGGCTTTTTGGGGGGTTTTCATTGCAGGATG |
6236 |
0.10061871798587015 |
No Hit |
| TAAGCAATGCCACAGGGGCATGATATCCTGGCTTCTTCTGACCAAGCAAA |
6223 |
0.10040896119725302 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
6223 |
0.10040896119725302 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
6217 |
0.10031215037173744 |
No Hit |
| AGCCCACCCACAGACCCACCTTCCAGCAGATCTGCTCCTTCCTTCAGGAG |
6210 |
0.1001992044086359 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality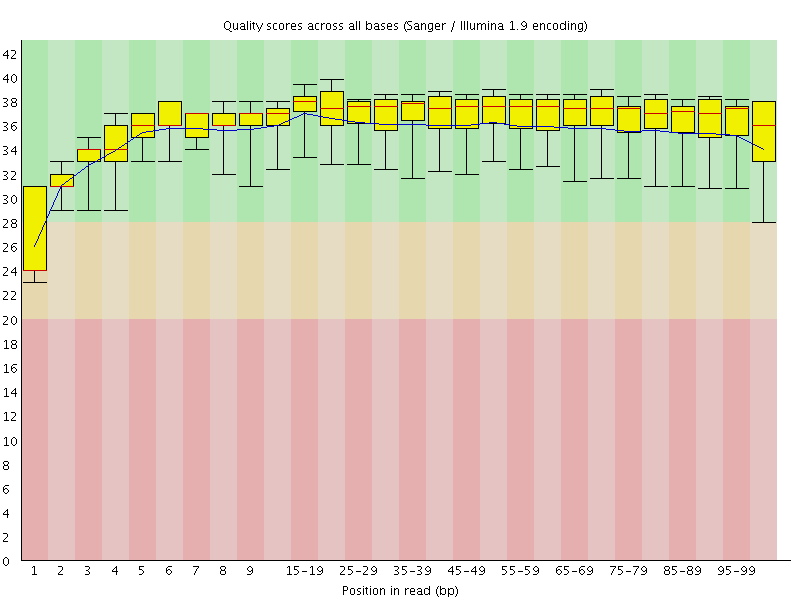
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores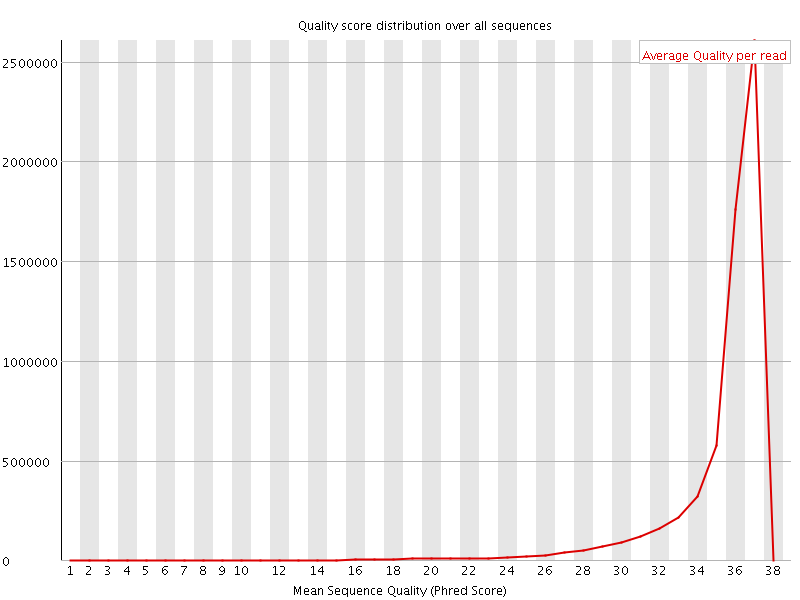
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content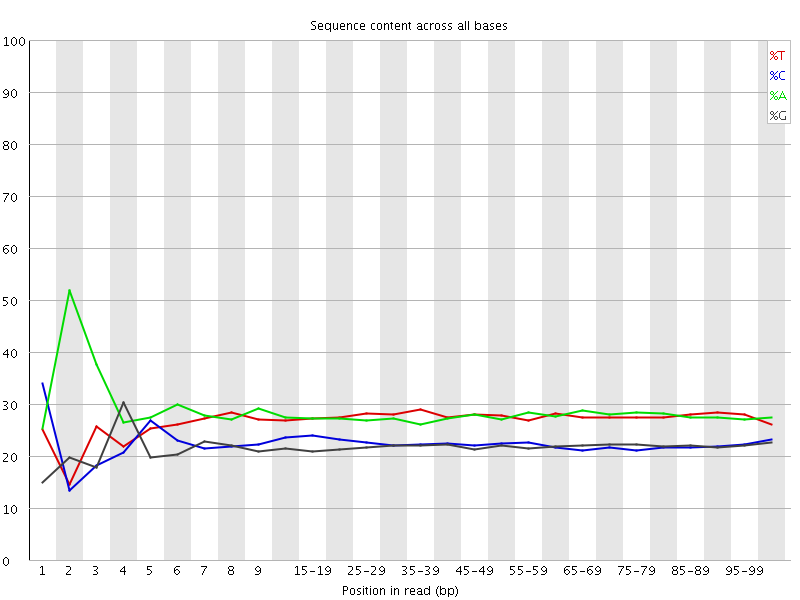
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content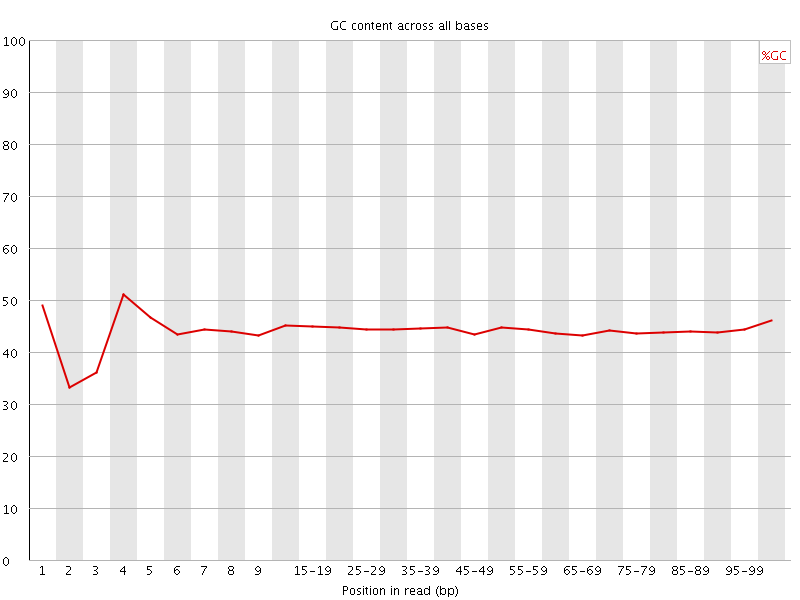
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content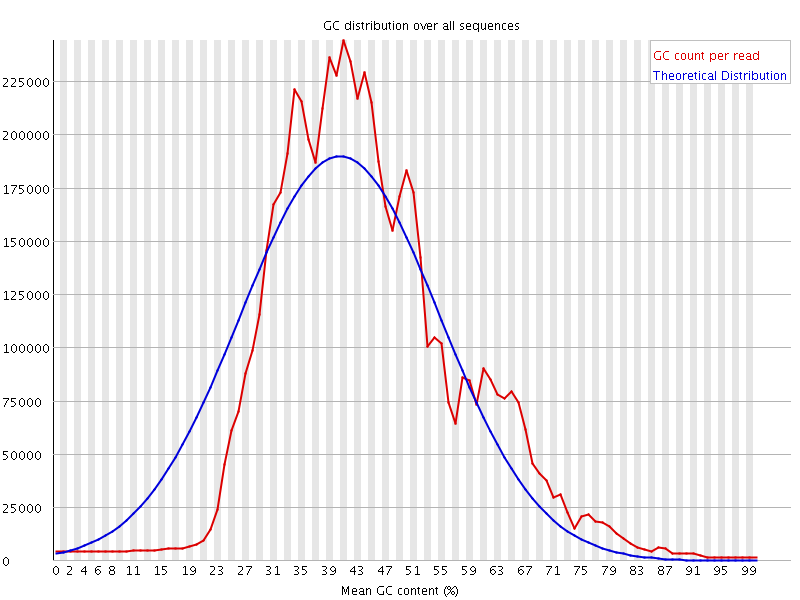
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution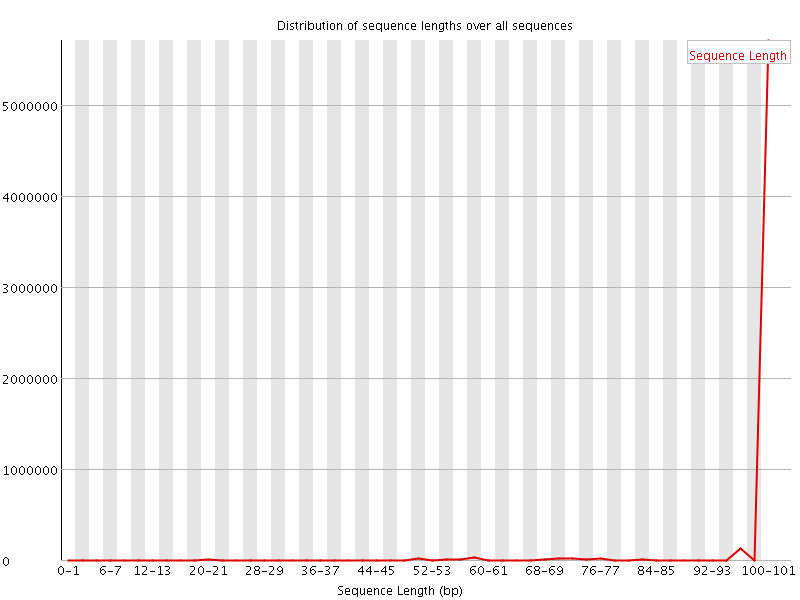
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels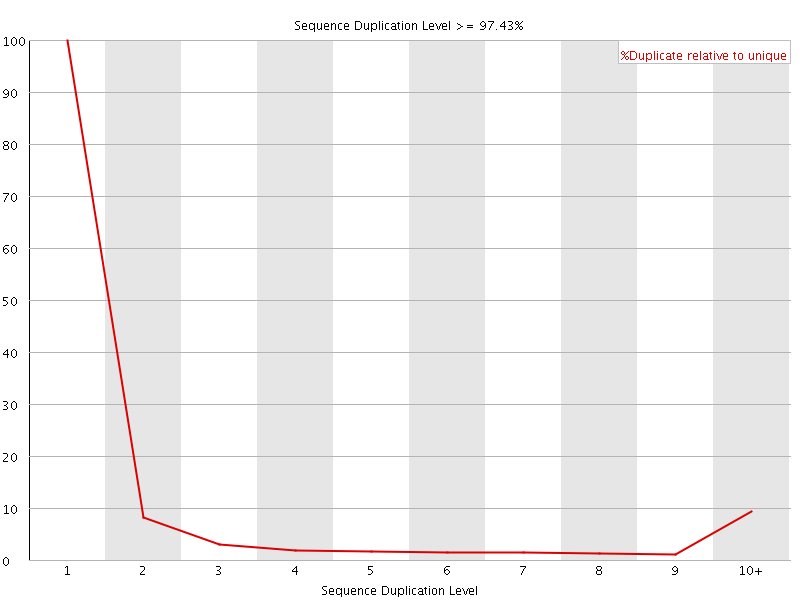
![[WARN]](Icons/warning.png) Overrepresented sequences
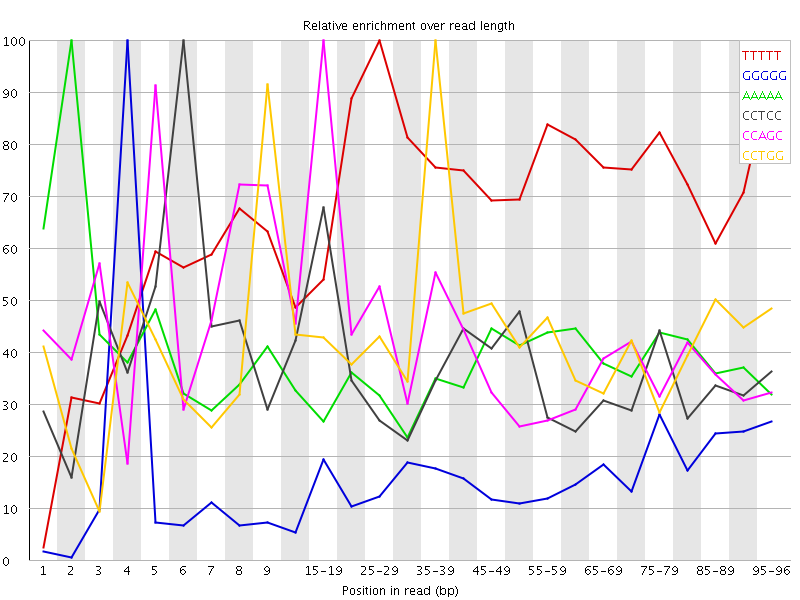
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content