| Sequence |
Count |
Percentage |
Possible Source |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
9489 |
0.24498957716384226 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
9397 |
0.2426142961965039 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8096 |
0.20902472512577372 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
8092 |
0.20892145204023724 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
8053 |
0.2079145394562569 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
7950 |
0.2052552575036933 |
No Hit |
| * |
6965 |
0.17982426019034262 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6894 |
0.17799116292207065 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6869 |
0.17734570613746783 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
6722 |
0.17355042024400333 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
6679 |
0.17244023457448648 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
6602 |
0.17045222767790982 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
6599 |
0.1703747728637575 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
6586 |
0.17003913533576404 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
6569 |
0.16960022472223413 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
6287 |
0.16231947219191445 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
6198 |
0.16002164603872843 |
No Hit |
| CAGGCTGGGTCCCAGCATGGCCTCAGCCTTCCTTCGCAGAAAGTTGAGCA |
6067 |
0.1566394524874097 |
No Hit |
| AGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTCTAGGCCCTGT |
6014 |
0.15527108410405177 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
6010 |
0.15516781101851532 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
5978 |
0.1543416263342237 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
5950 |
0.1536187147354686 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
5888 |
0.1520179819096536 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
5835 |
0.15064961352629566 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
5781 |
0.14925542687155358 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
5698 |
0.14711251034667225 |
No Hit |
| AGCCACCTCACCCACCCGGAATCTACATTTTCAATCAGTAAAAGTCACCA |
5609 |
0.14481468419348625 |
No Hit |
| CCGATGCCGATGCCCGAGGTGACCGGCGTCGGGGAGTAGGTGAAGGCGCCTGGATAGTGCATG |
5515 |
0.1423877666833797 |
No Hit |
| CATGTCATCTCAAAGGTCCCCAGGATTCGAACACCCAGTTATTCTCCAAC |
5491 |
0.141768128170161 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
5449 |
0.14068376077202827 |
No Hit |
| CGAGACCGCGCCCCAGGCCCAGCACTCACAAATTCCTGGTCGTGGTTATT |
5425 |
0.14006412225880957 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
5361 |
0.1384117528902264 |
No Hit |
| GGCACAAGGGTACCTACGGGCTGCTGCGGCGGCGAGAGGACTGGCCCTCC |
5343 |
0.13794702400531236 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
5267 |
0.13598483538011982 |
No Hit |
| TGCTCTGTGGTCCGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTG |
5248 |
0.1354942882238217 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
5241 |
0.13531356032413291 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
5234 |
0.13513283242444413 |
No Hit |
| CCGCGGCGCAGATCGAAACAGATCGGGCGGCTCGGGTTACACACGCACGC |
5183 |
0.13381610058385437 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
5034 |
0.12996917814762163 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
5007 |
0.1292720848202506 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
4907 |
0.12669025768183936 |
No Hit |
| CGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTGCGTGGCAGGATG |
4553 |
0.1175505896118636 |
No Hit |
| TCGTCTTCTGGGATGGCAGCTTTGCTTCAAGCGTCTAGTCTGTTAAAAGG |
4549 |
0.11744731652632714 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
4525 |
0.11682767801310845 |
No Hit |
| GAGGCACGAACACCCACGAGCGCCGCGTAACCGCAGCAGGTGGAGCGGGC |
4522 |
0.11675022319895612 |
No Hit |
| AGGGTCAGCCCACACCACCCAGCCCCCACGCCCAACCCTCGTGCCTCCCT |
4509 |
0.11641458567096265 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
4500 |
0.11618222122850563 |
No Hit |
| CATGCCCCTCCGCTGTCCCTTCGAGTCAGGTGTCTGTCCTTGGCTCCCAG |
4480 |
0.11566585580082339 |
No Hit |
| CATGGGGGCCAGTTGTGGGTGGTGGCCCAGGTGCAGGAGAGGCGGGCAGT |
4474 |
0.11551094617251872 |
No Hit |
| CGCAGTGACGCCCCCAATCTGCGCGCGGGACCAGCACACCAACTTTATCG |
4467 |
0.11533021827282994 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
4459 |
0.11512367210175704 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
4371 |
0.11285166421995516 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
4315 |
0.11140584102244486 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
4302 |
0.11107020349445139 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
4300 |
0.11101856695168318 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
4259 |
0.10996001782493457 |
No Hit |
| GGCTCCGCAGCCCCTGCCGCCGCCGCCGCCGCCTTCACCGCCGCCGCGTT |
4197 |
0.10835928499911959 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
4130 |
0.10662946081638407 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
4103 |
0.10593236748901304 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
4031 |
0.10407345194935694 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
4026 |
0.10394436059243638 |
No Hit |
| GCCGCTCACGGCACAGCCCAGTAAAGATAAGAGGCCTTCCATCACCGGTA |
4022 |
0.10384108750689992 |
No Hit |
| TGAGACCCGAAGACAGGGCCCACACCTCCAGCCCTTCTAAAACATAGTTA |
4017 |
0.10371199614997936 |
No Hit |
| CAGCAGTCCCAGCAGCCCCAGCCTCTACAGAAGCAGCCACCACAGCCCCA |
4015 |
0.10366035960721115 |
No Hit |
| CATGATATCTCGAGCCAATCCAAAGTCACATATCTTCACCACTTTCCCGT |
3999 |
0.10324726726506535 |
No Hit |
| GCTGCTACGTCCCTAATCCCCCTTGTTGGGAGGATTTTCGTATCACCCTT |
3984 |
0.10285999319430367 |
No Hit |
| GCAGAGCAGCTAGTCATTCAGCAGAGGCCGGCCAGAGAGGGCCCGGCTCA |
3966 |
0.10239526430938964 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
3963 |
0.1023178094952373 |
No Hit |
| CATGCTCCTCCTTGGCTCATCTTCAAACCGTCTCCTGTTTTGTAGTCCAA |
3962 |
0.10229199122385318 |
No Hit |
| CATGTACAAGGCCTTGCTACCTCAGCAGTCCTACAGCTTGGCCCAGCCGC |
3936 |
0.10162071616786628 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
3923 |
0.10128507863987281 |
No Hit |
| AGCACCTCACGGCCCCCACAGCAGATGGTGCAGTAGGACTGGTAGCCGTC |
3904 |
0.10079453148357467 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
3890 |
0.1004330756841971 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality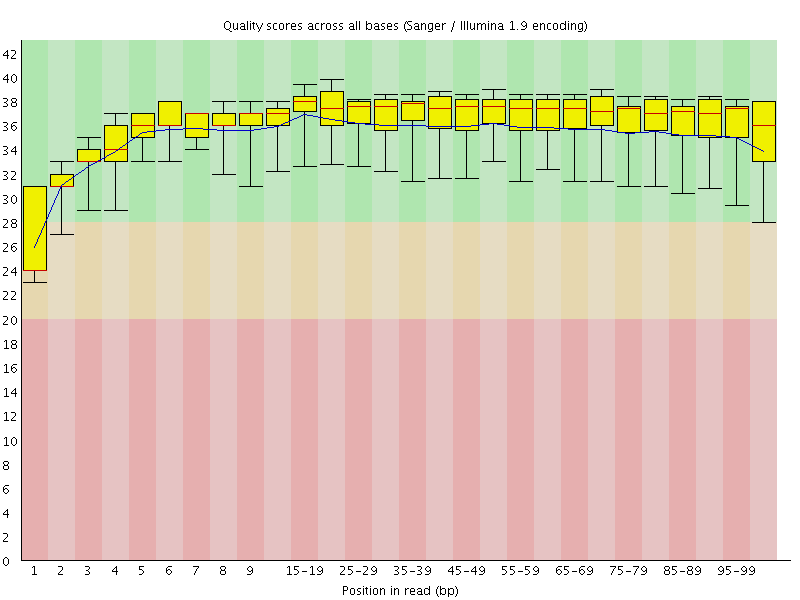
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores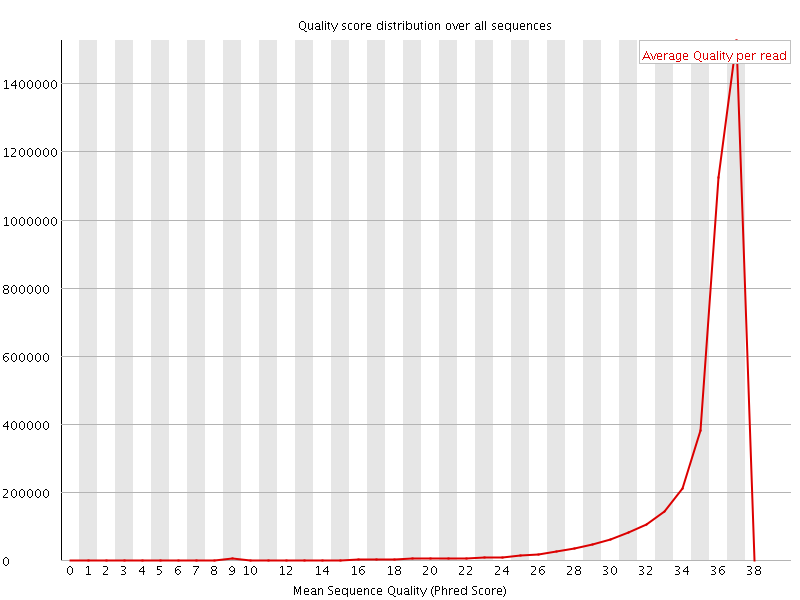
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content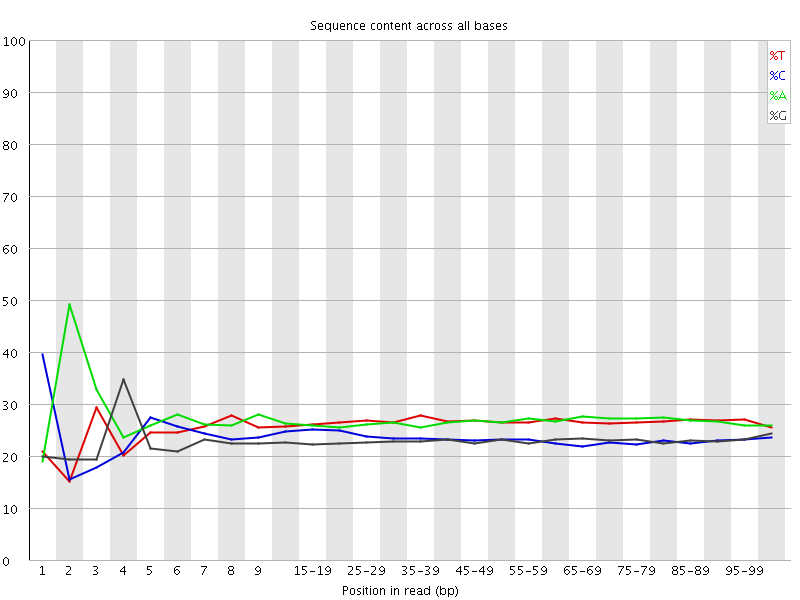
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content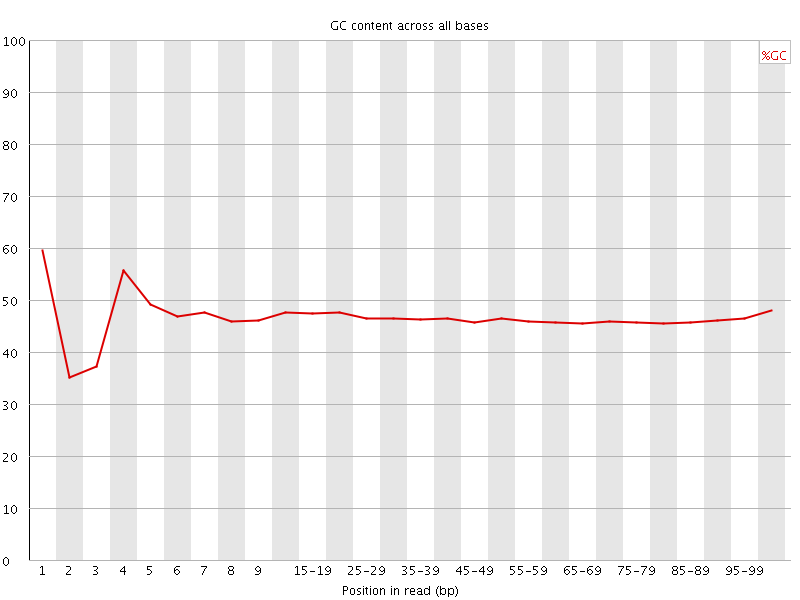
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content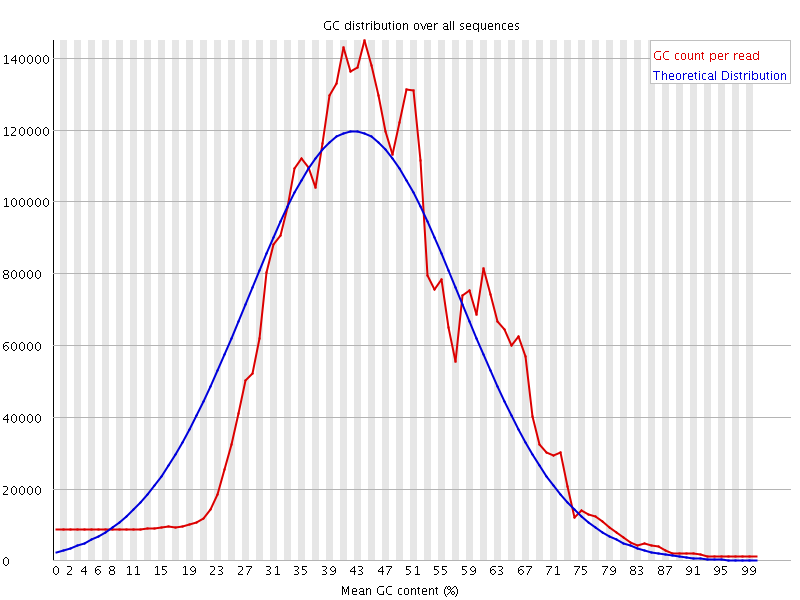
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution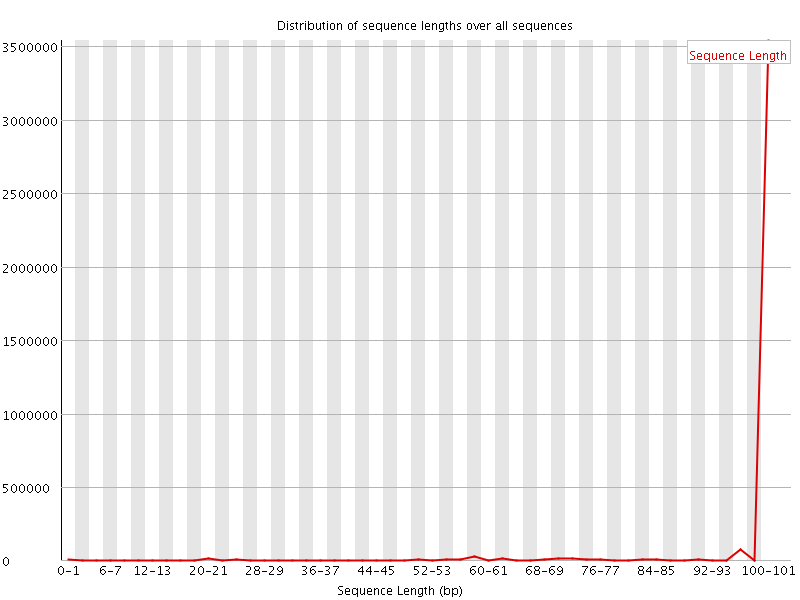
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels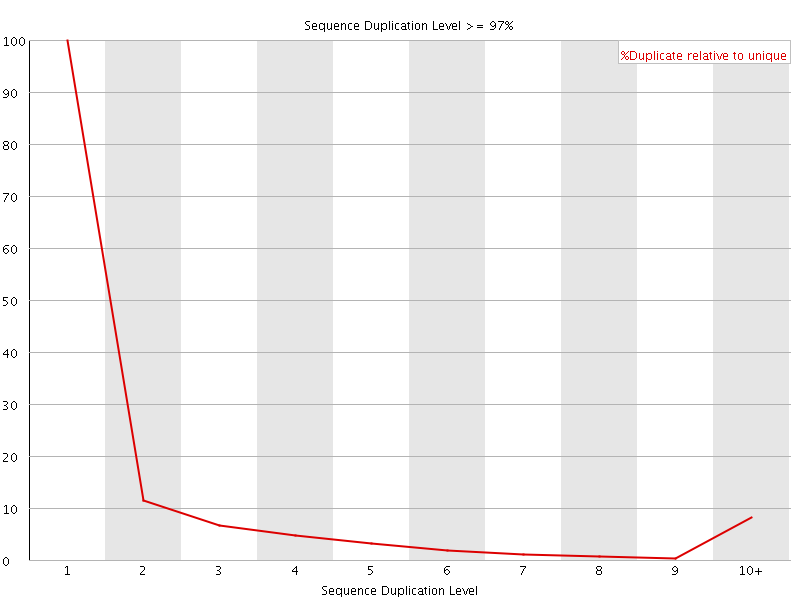
![[WARN]](Icons/warning.png) Overrepresented sequences
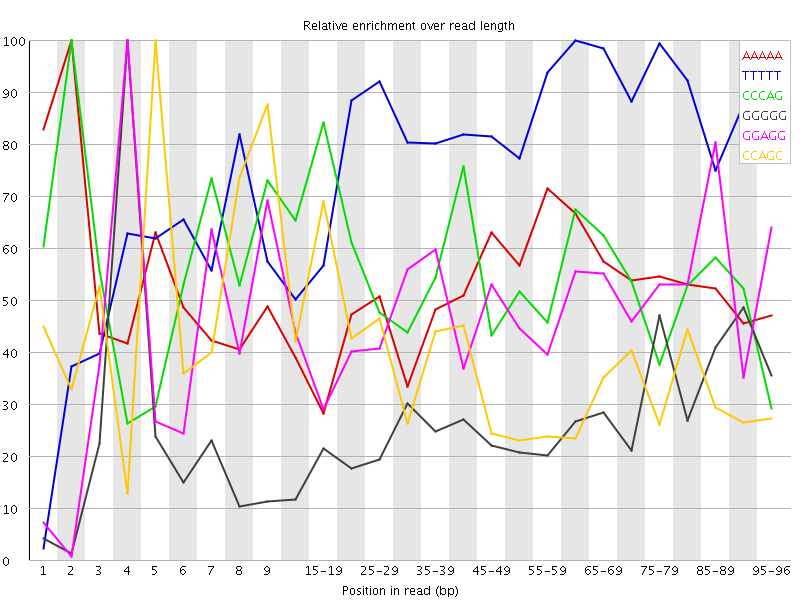
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content