| Sequence |
Count |
Percentage |
Possible Source |
| TGTGTCGCTGGTGCGGTCTCACTGCAGCCTGCAGAGCCACA |
20364 |
0.40559758123304535 |
No Hit |
| TGTGGCTCTGCAGGCTGCAGTGAGACCGCACCAGCGACACA |
19818 |
0.3947226902807156 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCAGCTTGGCGCAATCATGACACA |
14739 |
0.29356230356481317 |
No Hit |
| TGTGTCATGATTGCGCCAAGCTGAGACCGCACCAGCGACACA |
14457 |
0.2879456016443791 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCACGAGACTGCCCTTAACCTACACA |
13124 |
0.26139573050984516 |
No Hit |
| TGTGTAGGTTAAGGGCAGTCTCGTGAGACCGCACCAGCGACACA |
12927 |
0.25747200611862 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
12584 |
0.2506403438536949 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
12573 |
0.25042125264403253 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
11978 |
0.23857040993957068 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
11917 |
0.2373554495950796 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
9100 |
0.18124818253882893 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
9045 |
0.18015272649051733 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
8362 |
0.16654915410875687 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
8345 |
0.16621055860291511 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
8300 |
0.16531427638156926 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
8238 |
0.16407939865438162 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
8199 |
0.1633026207292152 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
8102 |
0.16137063460764747 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
8041 |
0.1601556742631564 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
7685 |
0.15306508602317587 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
7626 |
0.151889960444078 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
7568 |
0.15073475224767663 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
7351 |
0.14641268020251996 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
7301 |
0.14541681106769122 |
No Hit |
| * |
7254 |
0.1444806940809522 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
7009 |
0.13960093532029144 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7004 |
0.13950134840680856 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
6783 |
0.13509960683086558 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
6735 |
0.13414357246143 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
6695 |
0.133346877153567 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATGTGGGGTTTTTTT |
6682 |
0.13308795117851152 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
6663 |
0.1327095209072766 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
6626 |
0.13197257774750334 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
6580 |
0.13105637814346094 |
No Hit |
| ATACTTGTAATTCCCCTAACCATAAGATTTACTGCTGCTGTGGATATCTC |
6513 |
0.12972191350279044 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
6512 |
0.12970199612009384 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
6394 |
0.12735174496189805 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
6386 |
0.12719240590032546 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
6362 |
0.12671438871560767 |
No Hit |
| CATGACTATAAATATGAAGCAAATATGATTATCCTCACAAGTTCTGGAAT |
6304 |
0.12555918051920634 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
6296 |
0.12539984145763372 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
6253 |
0.12454339400168103 |
No Hit |
| GTATCTTAATGATGTATATAATTGCCTTCAATCCCCTTCTCACCCCACCC |
6249 |
0.12446372447089474 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACATTTTAGAGATATCTGGGTTTTTTTC |
6109 |
0.1216752908933743 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
5820 |
0.11591916729406422 |
No Hit |
| CATGACTATAAATATCAAGCAAATATGATTTTACTCACAAGTTCTGGAAT |
5785 |
0.11522205889968411 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATCTGGGTTTTTTTC |
5784 |
0.11520214151698753 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
5725 |
0.11402701593788962 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
5700 |
0.11352908137047527 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
5689 |
0.11330999016081296 |
No Hit |
| TGATCCCTTTTTCCTTTTCTCTTTCCTTGACTCCCGCTTATTCTCCTTTT |
5618 |
0.11189585598935615 |
No Hit |
| GACTGAAAGAGGGAGAAATAAAGACAAGGCCCCCGAGGAGCTGTCCAAAG |
5571 |
0.11095973900261714 |
No Hit |
| ATGTCAGATATAAAGAACTCACCGAACAGCAGCTCCCAGGCGCACTTCCT |
5568 |
0.11089998685452741 |
No Hit |
| TAAAACATCGCCTACAGAAAAGCGTATGAAAGGAGTGTAAGCTTTGCTCT |
5548 |
0.11050163920059593 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
5522 |
0.10998378725048498 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
5509 |
0.1097248612754295 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
5441 |
0.10837047925206245 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
5429 |
0.10813147065970355 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
5426 |
0.10807171851161383 |
No Hit |
| CAATTCAAATAAAAACGAAACTGTGTCAATTAGTTGAAGTAATGATGGCA |
5411 |
0.1077729577711652 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
5411 |
0.1077729577711652 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
5379 |
0.10713560152487481 |
No Hit |
| CTGAGGAAATAAGATGTAGGGCATTTCTGATGTGACTCAGTGGGAAAACT |
5339 |
0.10633890621701184 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
5326 |
0.10607998024195636 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
5303 |
0.10562188043993515 |
No Hit |
| TAATTATGGAATATTTACCATATGGAAGTTTACGAGACTATCTTCAAAAA |
5292 |
0.10540278923027284 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
5283 |
0.10522353278600366 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
5239 |
0.10434716794735437 |
No Hit |
| CATGTCCTCCTTTCTACCAATAACCGCATATCTACTCTTTTTCTGTACTT |
5172 |
0.10301270330668388 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
5165 |
0.10287328162780786 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality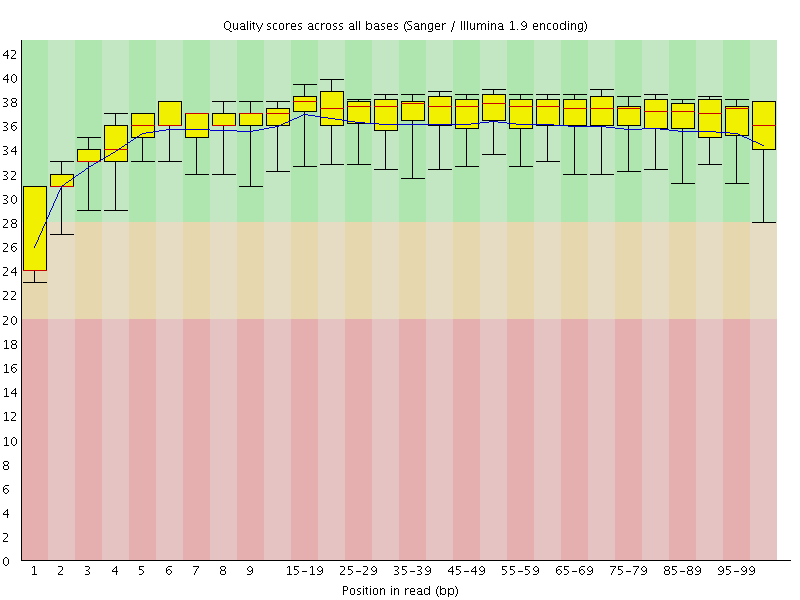
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores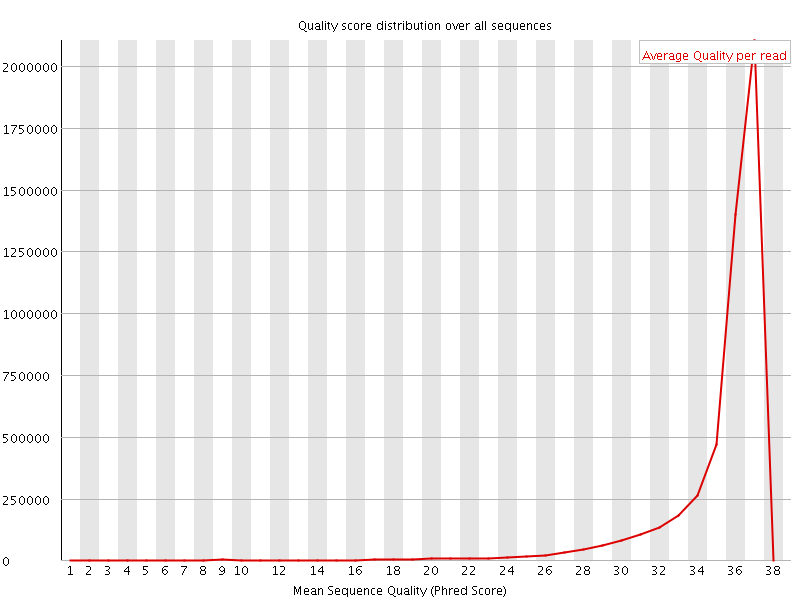
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content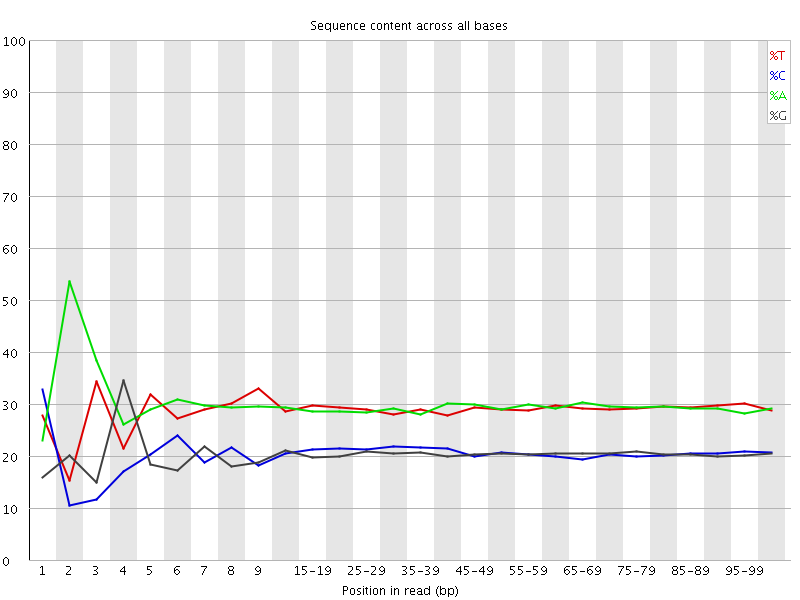
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content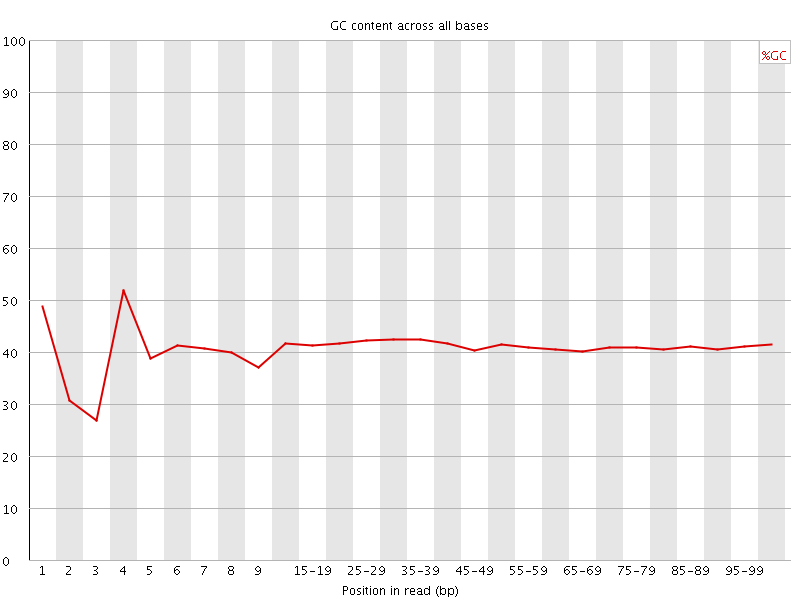
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content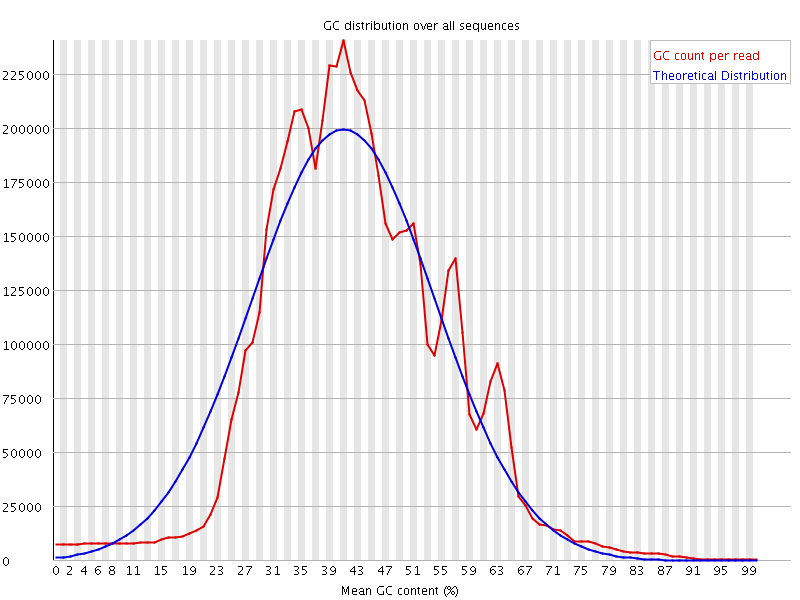
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution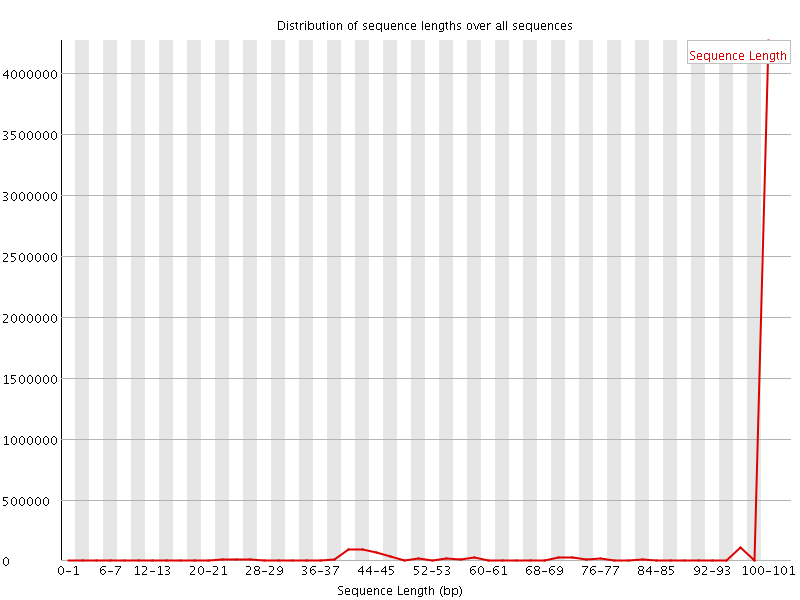
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels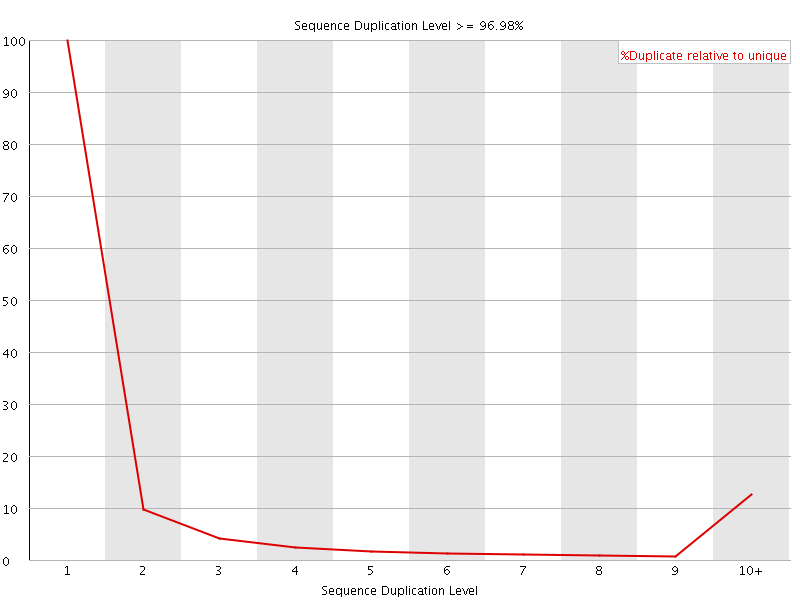
![[WARN]](Icons/warning.png) Overrepresented sequences
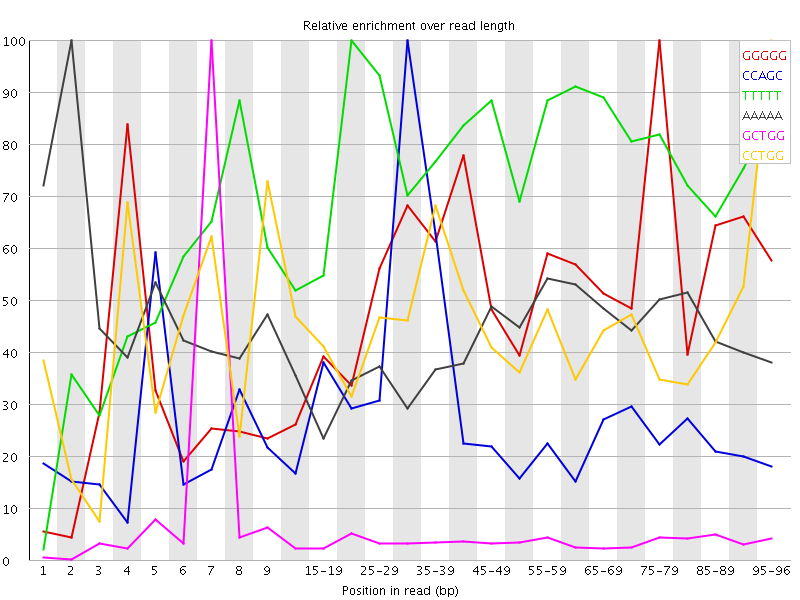
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content