| Sequence |
Count |
Percentage |
Possible Source |
| * |
17824 |
0.42873630071579527 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
11480 |
0.276138506071439 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
11427 |
0.274863650599158 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
8407 |
0.2022209425559746 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
8370 |
0.2013309491130614 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
8360 |
0.20109041034470648 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
8280 |
0.1991661001978672 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
7511 |
0.18066866891137445 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
7489 |
0.18013948362099363 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
7471 |
0.1797065138379548 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
7069 |
0.17003685535008733 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6684 |
0.1607761127684232 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
6644 |
0.1598139576950036 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6642 |
0.15976584994133258 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6640 |
0.1597177421876616 |
No Hit |
| AATCTCTTTCCTTTATAAATTACCCAGTCTTGCATATGTCTTTATTAGCA |
6577 |
0.15820234794702565 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
6573 |
0.1581061324396837 |
No Hit |
| ATACTTGTAATTCCCCTAACCATAAGATTTACTGCTGCTGTGGATATCTC |
6499 |
0.15632614555385735 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
6430 |
0.15466642805220845 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
6417 |
0.1543537276533471 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6383 |
0.15353589584094035 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6364 |
0.15307887218106606 |
No Hit |
| CATGCCCACAGAGACTTGCACAACATGCAGAATGGCAGCACATTGGTAAG |
6302 |
0.15158753181726559 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
6178 |
0.14860485108966467 |
No Hit |
| CTGAGGAAATAAGATGTAGGGCATTTCTGATGTGACTCAGTGGGAAAACT |
6075 |
0.14612730177560906 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
5997 |
0.14425109938244077 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
5902 |
0.1419659810830691 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
5848 |
0.14066707173395257 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
5788 |
0.1392238391238231 |
No Hit |
| CGAGACCGCGCCCCAGGCCCAGCACTCACAAATTCCTGGTCGTGGTTATT |
5671 |
0.13640953553407065 |
No Hit |
| GGCACAAGGGTACCTACGGGCTGCTGCGGCGGCGAGAGGACTGGCCCTCC |
5601 |
0.13472576415558624 |
No Hit |
| TAACCAGAGTTAGTAGTGTATGTAAAATTCTGACTGTTCTTGCAACAATA |
5304 |
0.12758176273544536 |
No Hit |
| AGTTCTAACATTTTGTTTATGGCTTTTTGGGGGGTTTTCATTGCAGGATG |
5290 |
0.12724500845974848 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
5283 |
0.12707663132190003 |
No Hit |
| TAAGCAATGCCACAGGGGCATGATATCCTGGCTTCTTCTGACCAAGCAAA |
5248 |
0.12623474563265785 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
5224 |
0.12565745258860608 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
5195 |
0.12495989016037681 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
5185 |
0.12471935139202191 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
5184 |
0.12469529751518642 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
5166 |
0.12426232773214757 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
5159 |
0.12409395059429915 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
5156 |
0.12402178896379265 |
No Hit |
| AGAACAGTTGCAAATAGACTACATAATATACGTGGGCAAAAAGGCAATTA |
5126 |
0.12330017265872793 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
5114 |
0.12301152613670202 |
No Hit |
| TAATTGCCTTTTTGCCCACGTATATTATGTAGTCTATTTGCAACTGTTCT |
5085 |
0.12231396370847279 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
4899 |
0.11783994261707142 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
4867 |
0.11707021855833571 |
No Hit |
| TAATAGGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGT |
4843 |
0.11649292551428392 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
4840 |
0.11642076388377744 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
4821 |
0.1159637402239031 |
No Hit |
| ATCATGTCCCCAGAAAATCCAGTTACCTCTTGGTCAGTCATCCGGAAGCA |
4821 |
0.1159637402239031 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
4792 |
0.11526617779567387 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
4772 |
0.11478510025896405 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
4680 |
0.11257214359009883 |
No Hit |
| TAAACATGTCTGCAGAAGAGCAGCTGTTGCCCTTGGCCGAAAACGAGCTG |
4642 |
0.11165809627035019 |
No Hit |
| ATCCCCTCTTGCCATCTGTTTTTCAGAGGACCTACTAACTATGAAGAGCC |
4587 |
0.11033513304439817 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
4562 |
0.1097337861235109 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
4550 |
0.10944513960148498 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
4528 |
0.10891595431110418 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
4485 |
0.10788163760717807 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
4466 |
0.10742461394730374 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
4401 |
0.1058611119529968 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
4381 |
0.10538003441628697 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
4377 |
0.10528381890894502 |
No Hit |
| CCGCGGCGCAGATCGAAACAGATCGGGCGGCTCGGGTTACACACGCACGC |
4355 |
0.10475463361856421 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
4355 |
0.10475463361856421 |
No Hit |
| TGCTCTGTGGTCCGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTG |
4343 |
0.1044659870965383 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
4325 |
0.10403301731349947 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
4323 |
0.10398490955982849 |
No Hit |
| AGAAAATTTACTTTGGAAACCTACTTTTCACAGAAGTCATTCAGAACACC |
4237 |
0.10191627615197625 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality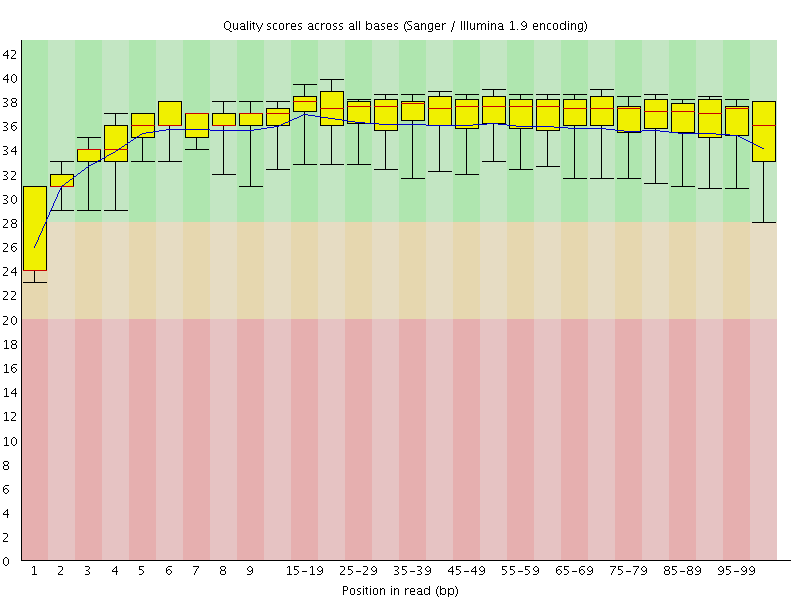
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores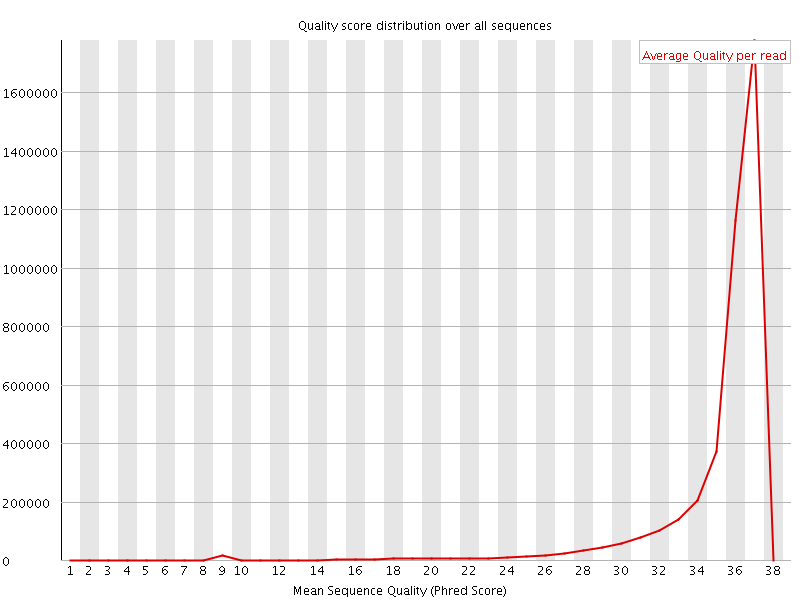
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content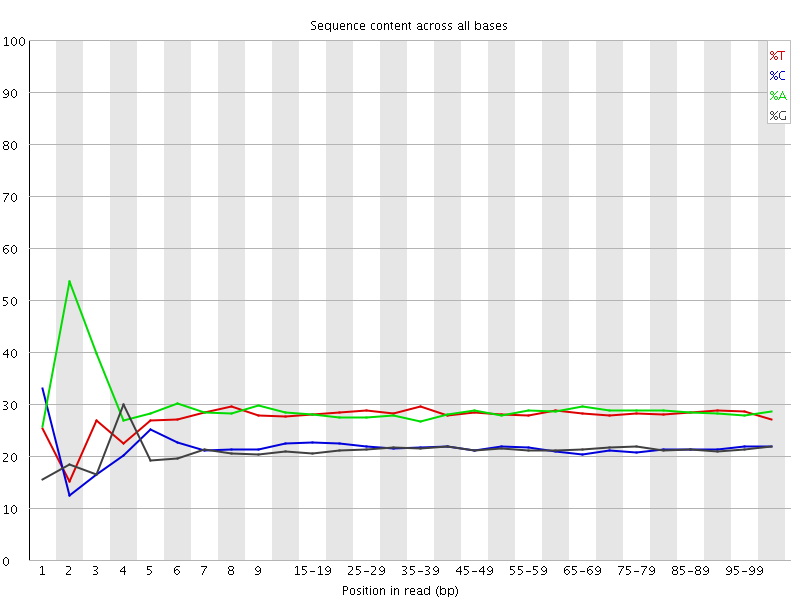
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content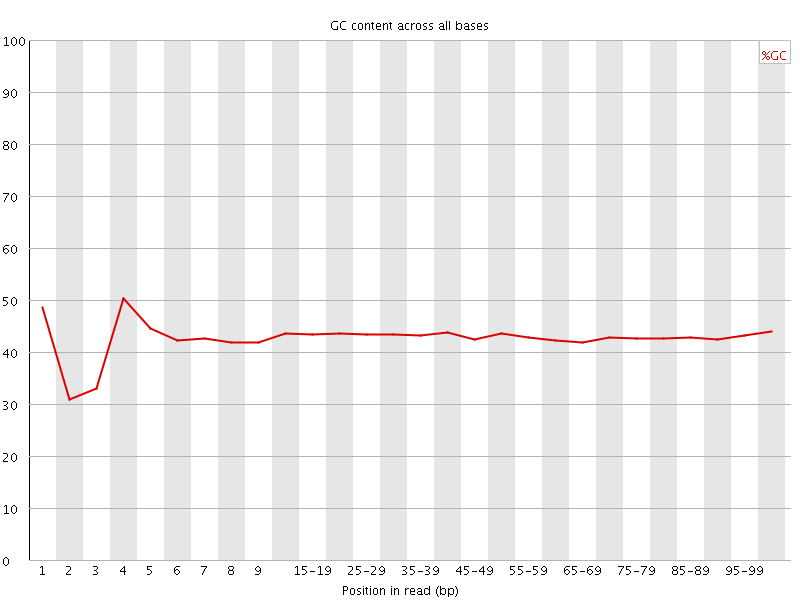
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content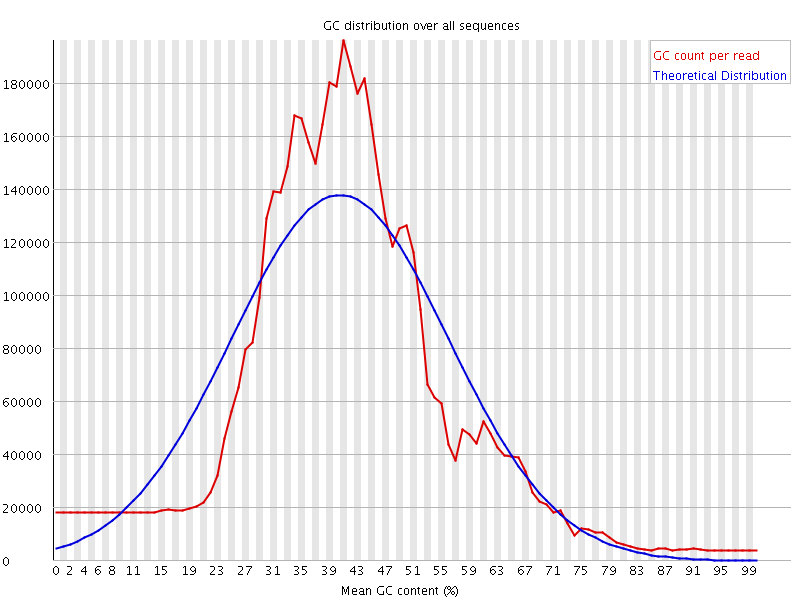
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution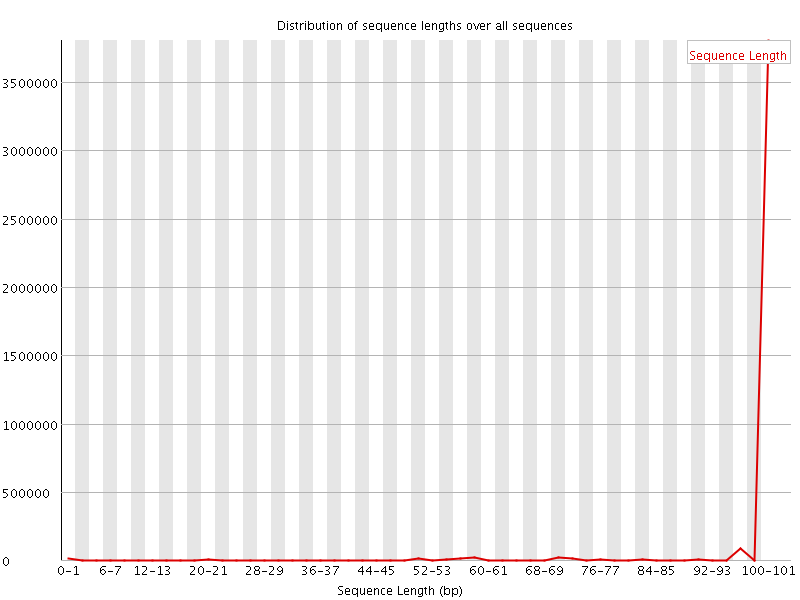
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels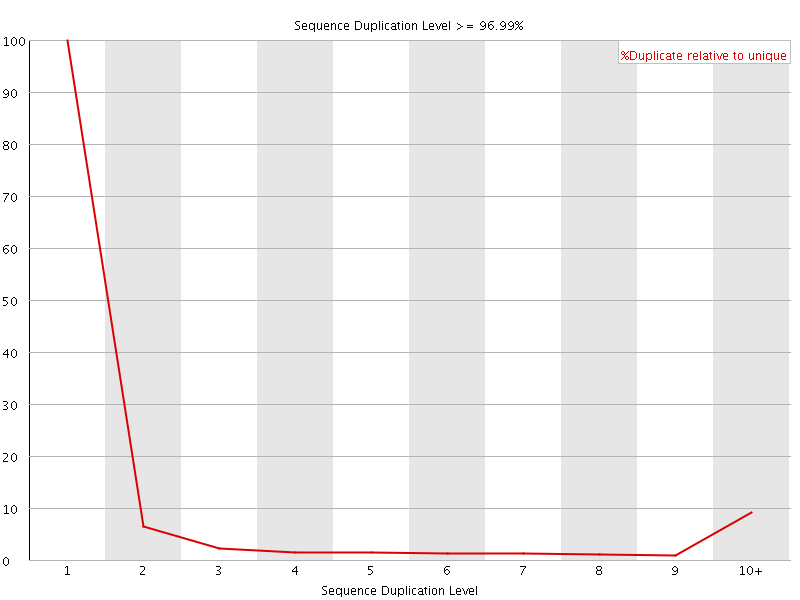
![[WARN]](Icons/warning.png) Overrepresented sequences
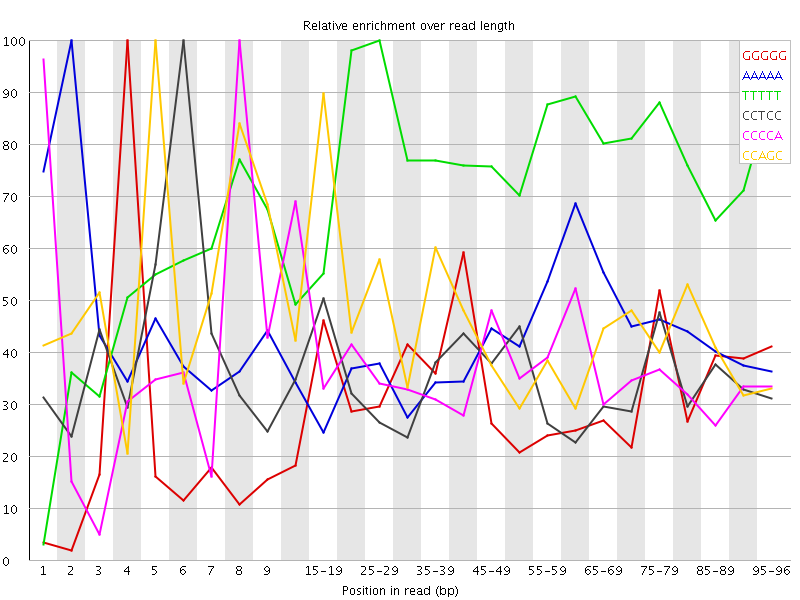
Overrepresented sequences![[FAIL]](Icons/error.png) Kmer Content
Kmer Content