| Sequence |
Count |
Percentage |
Possible Source |
| * |
11941 |
0.25123439594759617 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
10021 |
0.21083827835113148 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
9993 |
0.21024916830284968 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
9442 |
0.1986563241384476 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
9327 |
0.19623676501157603 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
8392 |
0.176564697327881 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
8350 |
0.17568103225545834 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8315 |
0.1749446446951061 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
8208 |
0.17269340272488645 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
8198 |
0.1724830062790715 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
7968 |
0.16764388802532837 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
7734 |
0.16272061119325923 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7702 |
0.1620473425666515 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
7655 |
0.16105847927132136 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7609 |
0.16009065562057273 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
7275 |
0.15306341433035442 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
7237 |
0.1522639078362577 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7160 |
0.1506438552034828 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
7114 |
0.14967603155273418 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
7068 |
0.14870820790198555 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
6961 |
0.1464569659317659 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
6942 |
0.14605721268471755 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
6898 |
0.14513146832313192 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
6829 |
0.14367973284700894 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
6746 |
0.14193344234674513 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
6736 |
0.1417230459009302 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
6719 |
0.14136537194304483 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
6702 |
0.14100769798515947 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
6667 |
0.14027131042480726 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
6662 |
0.1401661122018998 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
6637 |
0.1396401210873625 |
No Hit |
| GATCCAAAACTTCTCTCCGACACTGAATACCTTCTGCTTCTCCTATCATT |
6542 |
0.13764135485212076 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
6528 |
0.13734679982797987 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6447 |
0.135642588616879 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
6357 |
0.13374902060454474 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6339 |
0.1333703070020779 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
6241 |
0.13130842183309166 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6165 |
0.12970940884489826 |
No Hit |
| TAAACATGTCTGCAGAAGAGCAGCTGTTGCCCTTGGCCGAAAACGAGCTG |
6073 |
0.127773761543401 |
No Hit |
| ATCCCCTCTTGCCATCTGTTTTTCAGAGGACCTACTAACTATGAAGAGCC |
6071 |
0.12773168225423803 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
5991 |
0.12604851068771866 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
5968 |
0.12556459886234433 |
No Hit |
| ATCCTTCGAGGGCTTTGTCTGACAACTGTCTTATCTTCTTTTGTAAGTGG |
5942 |
0.12501756810322553 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
5936 |
0.12489133023573659 |
No Hit |
| TAATTCTGAACTGGAAAAGCCCCAGAAAGTCCGGAAAGACAAGGAAGGAA |
5903 |
0.12419702196454735 |
No Hit |
| TGATCCCTTTTTCCTTTTCTCTTTCCTTGACTCCCGCTTATTCTCCTTTT |
5877 |
0.12364999120542856 |
No Hit |
| GACTGAAAGAGGGAGAAATAAAGACAAGGCCCCCGAGGAGCTGTCCAAAG |
5870 |
0.12350271369335813 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
5776 |
0.12152498710269787 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
5717 |
0.12028364807238985 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
5656 |
0.11900022975291882 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
5654 |
0.11895815046375584 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
5549 |
0.1167489877826992 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5529 |
0.11632819489106935 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
5513 |
0.11599156057776547 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
5450 |
0.11466606296913148 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
5362 |
0.11281457424596017 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
5326 |
0.11205714704102647 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
5319 |
0.11190986952895603 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
5266 |
0.11079476836613694 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
5228 |
0.10999526187204026 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
5190 |
0.10919575537794354 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
5134 |
0.10801753528138 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
5114 |
0.10759674238975017 |
No Hit |
| AGAAGAGGGCTCCTTTTGTGCATATCAAACCTTGTTCCAGAATGGAGTGG |
5074 |
0.10675515660649047 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
5059 |
0.1064395619377681 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
5053 |
0.10631332407027914 |
No Hit |
| GACCCCCTCCACTAACTCCCGAGGACGTTGGCTTTGCATCTGGTTTTTCT |
5039 |
0.10601876904613824 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
5023 |
0.10568213473283439 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
5018 |
0.10557693650992692 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
4984 |
0.1048615885941562 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATGTGGGGTTTTTTA |
4955 |
0.10425143890129293 |
No Hit |
| GCCCTCCTCCTGCCACGGGCCTGCTCCCCTCCTTCTCTCATGGGGGTCTG |
4955 |
0.10425143890129293 |
No Hit |
| AATCTCTTTCCTTTATAAATTACCCAGTCTTGCATATGTCTTTATTAGCA |
4896 |
0.10301009987098489 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
4845 |
0.10193707799732882 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
4828 |
0.10157940403944345 |
No Hit |
| TGAGACCGCACCAGCGACACA |
4790 |
0.10077989754534675 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality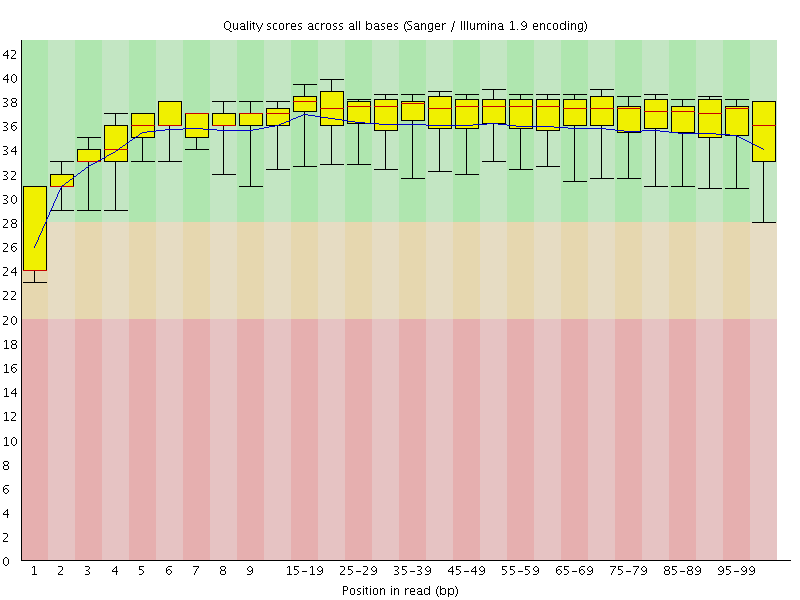
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores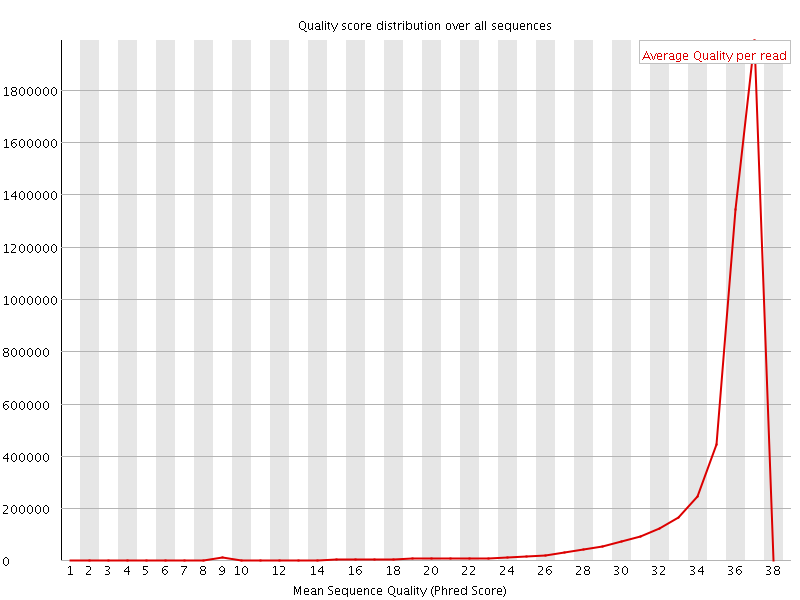
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content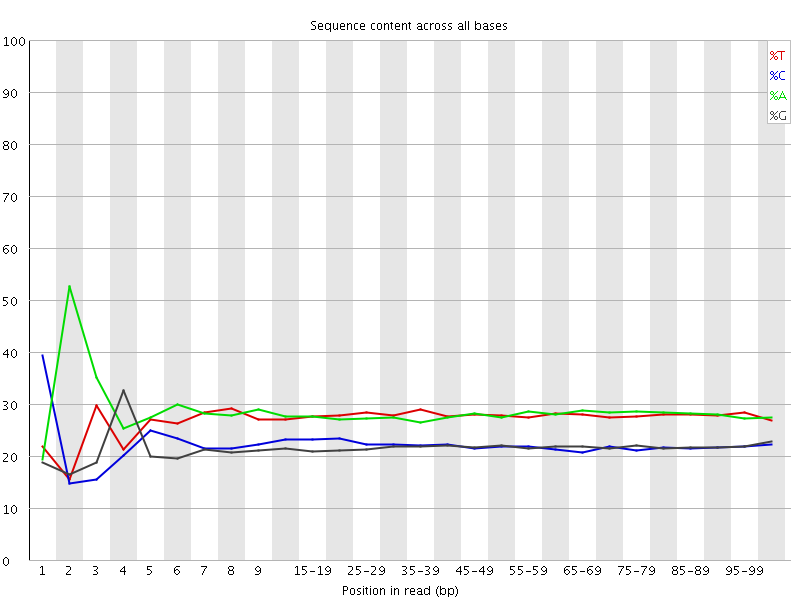
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content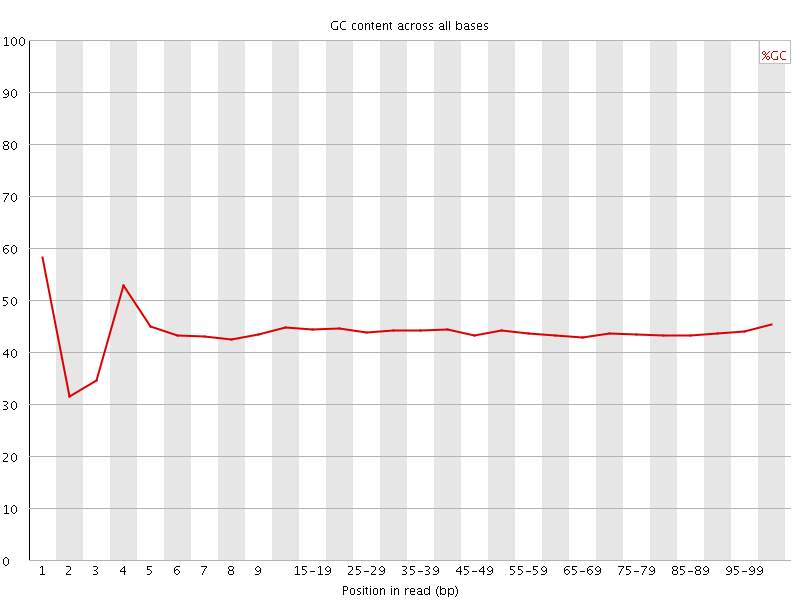
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content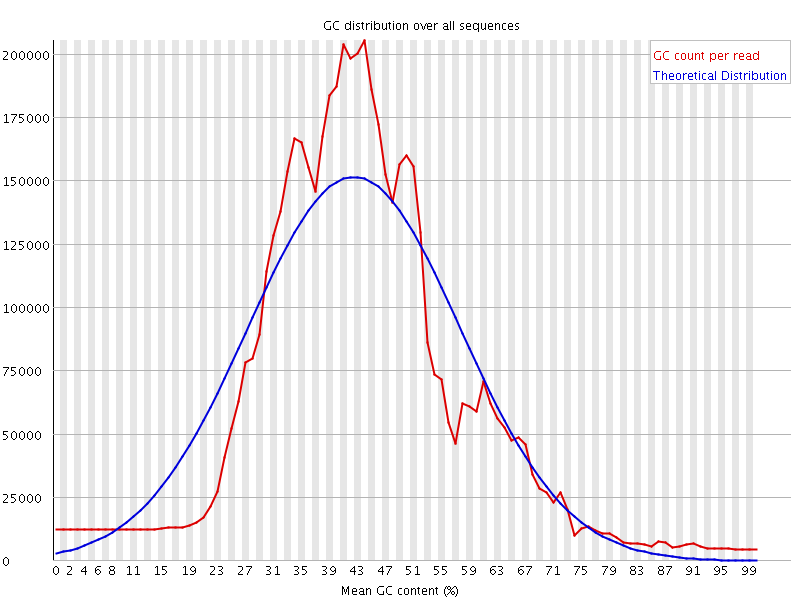
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution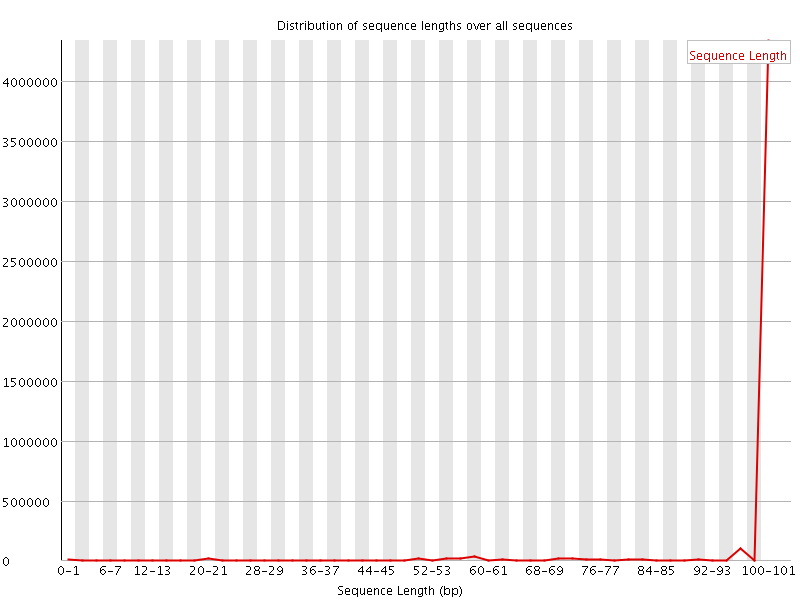
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels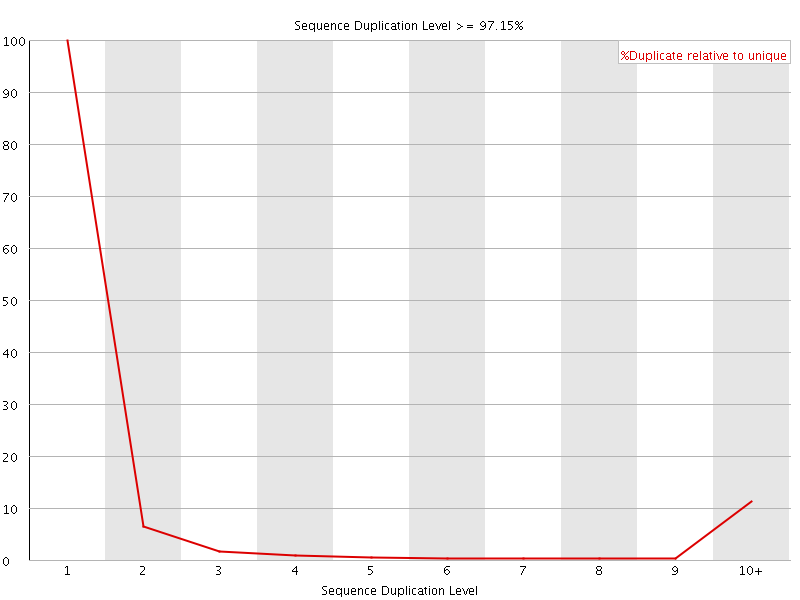
![[WARN]](Icons/warning.png) Overrepresented sequences
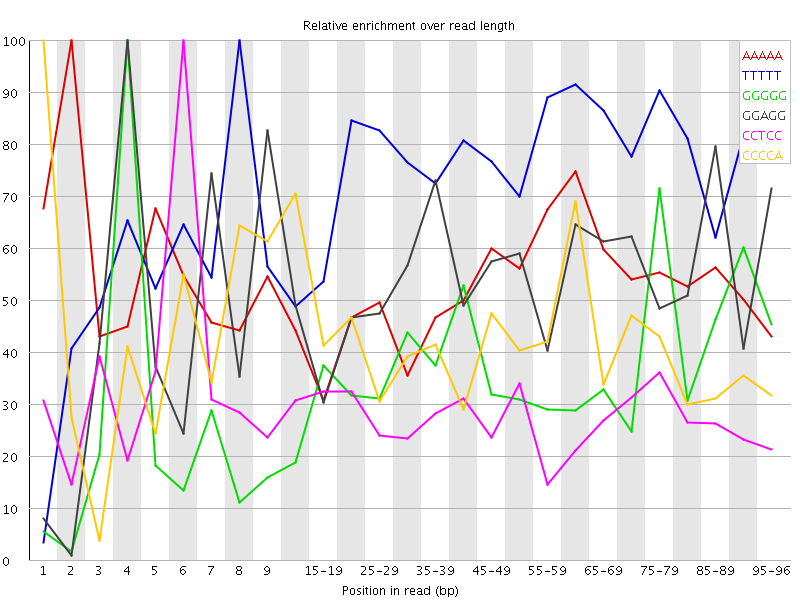
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content