| Sequence |
Count |
Percentage |
Possible Source |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
5431 |
0.23413337885807062 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
5422 |
0.23374538393821742 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
5420 |
0.23365916284491672 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
5403 |
0.23292628355186068 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
5391 |
0.23240895699205644 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
5353 |
0.23077075621934304 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
5053 |
0.2178375922242369 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
5012 |
0.21607005981157243 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
4537 |
0.19559255015265445 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
4514 |
0.1946010075796963 |
No Hit |
| * |
3817 |
0.1645529565643998 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
3709 |
0.15989701752616164 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
3674 |
0.15838814839339926 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
3663 |
0.15791393238024537 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
3624 |
0.15623262106088157 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
3511 |
0.1513611292893916 |
No Hit |
| TGAGACCGCACCAGCGACACA |
3507 |
0.1511886871027902 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
3489 |
0.15041269726308384 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
3476 |
0.14985226015662925 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
3460 |
0.14916249141022359 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
3449 |
0.1486882753970697 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
3442 |
0.14838650157051722 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
3342 |
0.14407544690548185 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
3323 |
0.14325634651912514 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
3269 |
0.14092837700000604 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
3251 |
0.14015238716029965 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
3236 |
0.13950572896054436 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
3200 |
0.13795374928113163 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
3187 |
0.13739331217467704 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
3169 |
0.13661732233497068 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
3155 |
0.13601377468186573 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
3129 |
0.13489290046895655 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
3125 |
0.13472045828235513 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
3101 |
0.1336858051627466 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
3100 |
0.13364269461609626 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
3080 |
0.1327804836830892 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
3055 |
0.13170272001683034 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
3025 |
0.13040940361731976 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
2950 |
0.12717611261854322 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
2912 |
0.1255379118458298 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
2903 |
0.1251499169259766 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
2877 |
0.12402904271306742 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
2831 |
0.12204595756715114 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
2815 |
0.12135618882074548 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
2743 |
0.11825222946192002 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
2741 |
0.11816600836861932 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
2731 |
0.11773490290211577 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
2670 |
0.11510515955644421 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
2569 |
0.11075099434475848 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
2557 |
0.11023366778495425 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
2519 |
0.10859546701224082 |
No Hit |
| GAGACCGCACCAGCGACACC |
2512 |
0.10829369318568834 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
2498 |
0.10769014553258338 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
2476 |
0.1067417135062756 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
2453 |
0.10575017093331747 |
No Hit |
| GGTGTCGCTGGTGCGGTCTC |
2444 |
0.10536217601346427 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
2439 |
0.10514662328021253 |
No Hit |
| GCCCTCCTCCTGCCACGGGCCTGCTCCCCTCCTTCTCTCATGGGGGTCTG |
2430 |
0.10475862836035935 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
2429 |
0.10471551781370898 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
2426 |
0.10458618617375792 |
No Hit |
| GCACGTGGGAGAGTAAGCAGGCCAGGTAGAGCTGCAGAAAACTAAGGCCC |
2410 |
0.10389641742735226 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
2408 |
0.10381019633405154 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
2385 |
0.10281865376109342 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
2349 |
0.10126667408168069 |
No Hit |
| CATGCACTATCCAGGCGCCTTCACCTACTCCCCGACGCCGGTCACCTCGGGCATCGGCATCGG |
2339 |
0.10083556861517715 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
2332 |
0.10053379478862468 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
2332 |
0.10053379478862468 |
No Hit |
| AGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTCTAGGCCCTGT |
2323 |
0.10014579986877148 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality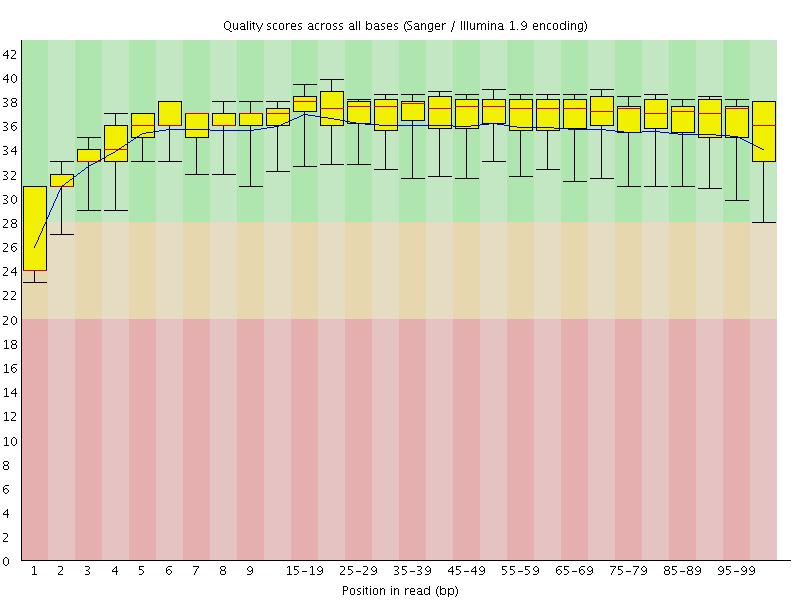
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores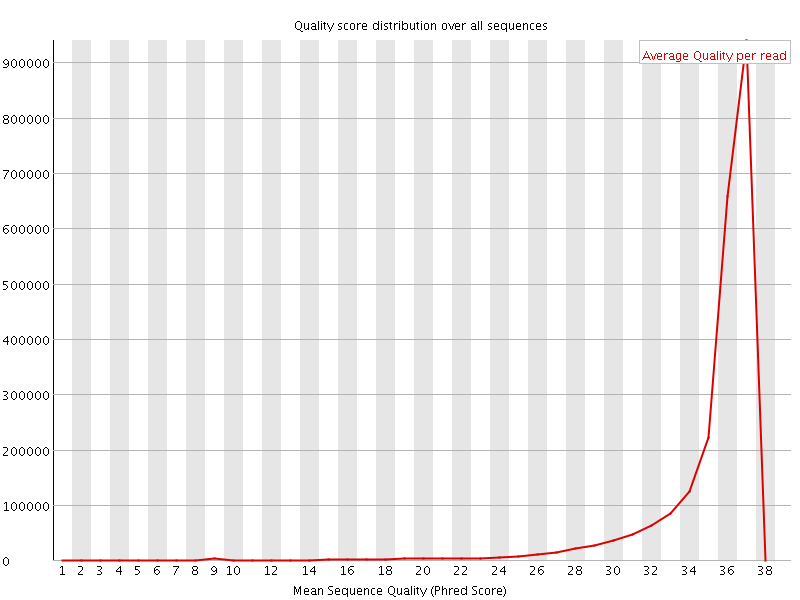
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content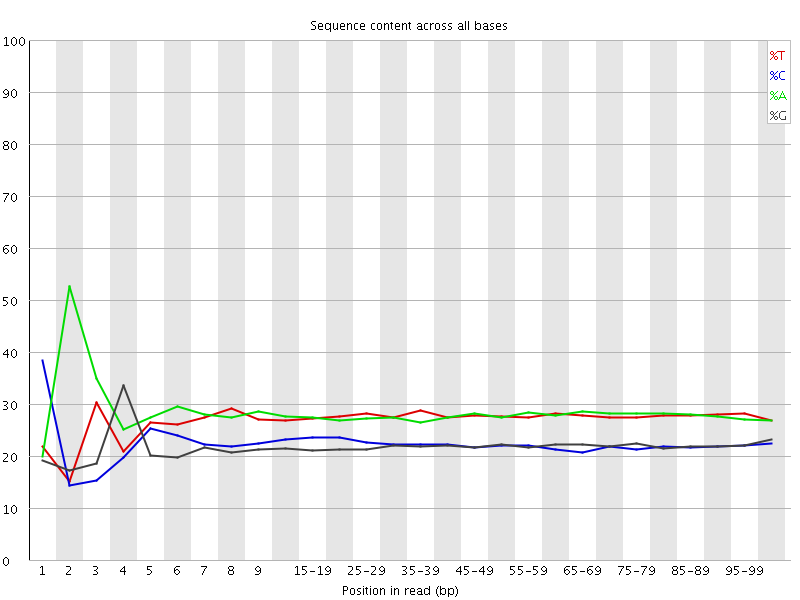
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content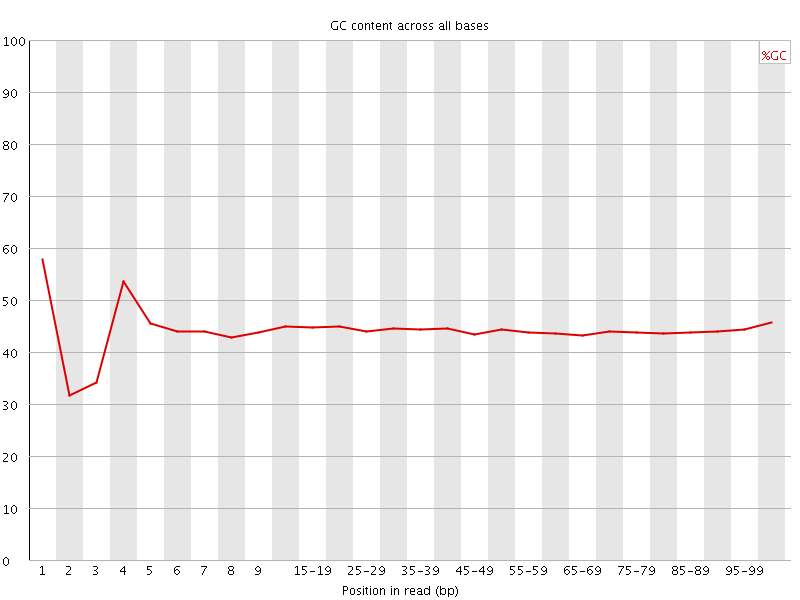
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content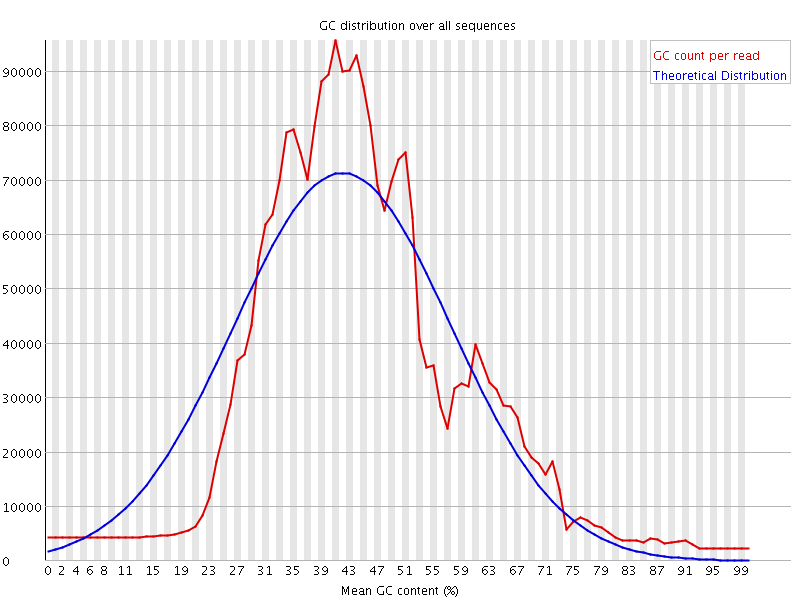
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution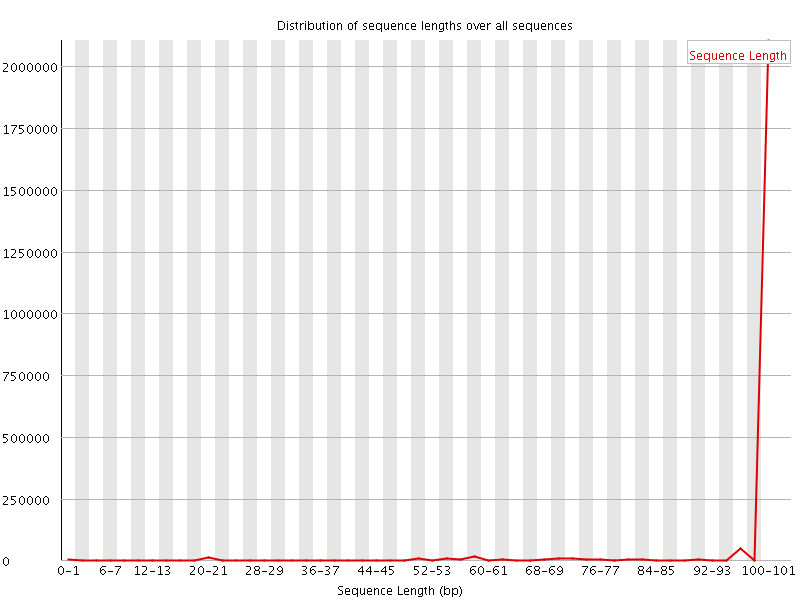
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels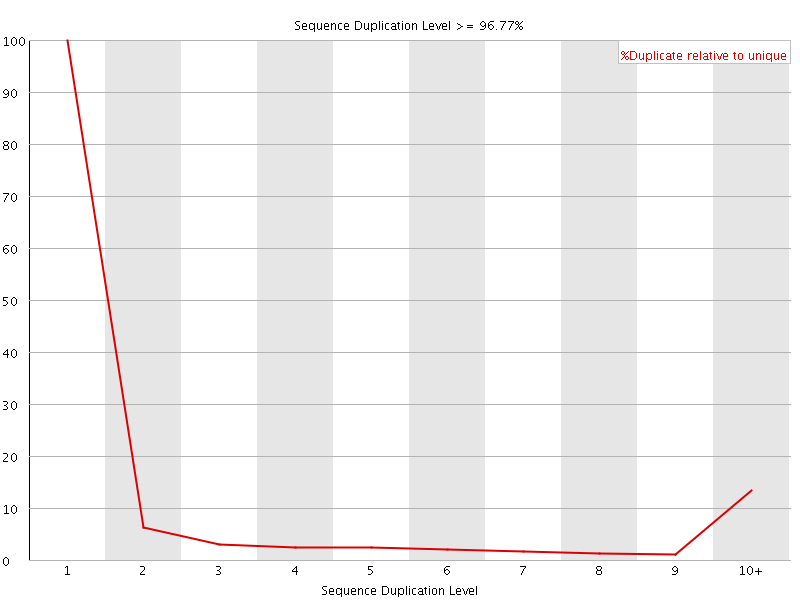
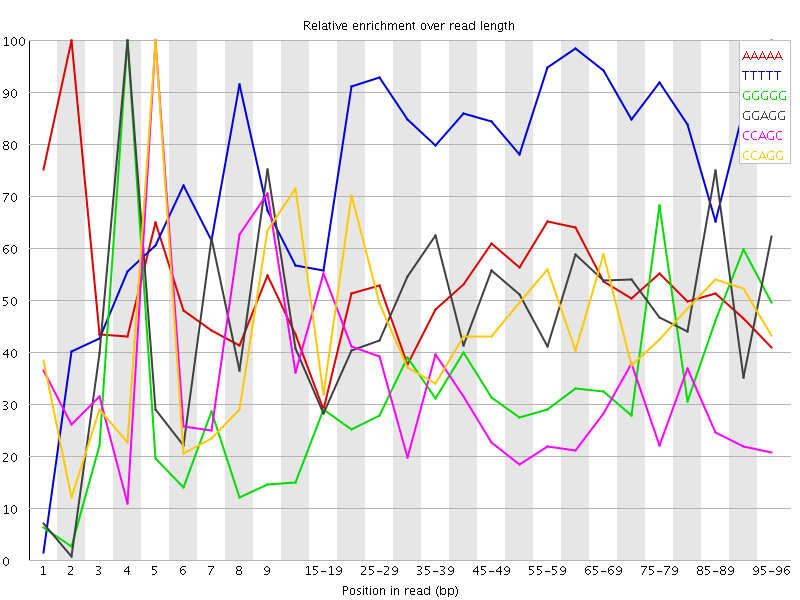
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content