| Sequence |
Count |
Percentage |
Possible Source |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
13903 |
0.2815836248478967 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
13824 |
0.2799836028121501 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
11050 |
0.22380055056960782 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
11034 |
0.22347649547376042 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
10459 |
0.21183076546674465 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
10379 |
0.2102104899875077 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
9847 |
0.19943565805058175 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
9826 |
0.19901033573728205 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCACGAGACTGCCCTTAACCTACACA |
8928 |
0.18082274348284696 |
No Hit |
| TGTGTAGGTTAAGGGCAGTCTCGTGAGACCGCACCAGCGACACA |
8795 |
0.17812903549861547 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
8383 |
0.16978461678054504 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
8309 |
0.16828586196225082 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
8255 |
0.16719217601376585 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
8178 |
0.16563266086500025 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
8071 |
0.1634655424115208 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
8025 |
0.16253388401095956 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
7979 |
0.16160222561039828 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
7693 |
0.1558097407721261 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
7633 |
0.15459453416269833 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
7631 |
0.15455402727571743 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
7489 |
0.1516780383000718 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
7474 |
0.15137423664771485 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
7411 |
0.15009826970781573 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
7348 |
0.14882230276791658 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
7329 |
0.1484374873415978 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7279 |
0.1474248151670747 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
7007 |
0.14191587853766896 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
6989 |
0.14155131655484066 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
6853 |
0.13879684824013777 |
No Hit |
| CAATTCAAATAAAAACGAAACTGTGTCAATTAGTTGAAGTAATGATGGCA |
6583 |
0.13332841849771299 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
6528 |
0.13221447910573755 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
6524 |
0.1321334653317757 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
6497 |
0.1315866223575332 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
6493 |
0.13150560858357138 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
6490 |
0.1314448482531 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6356 |
0.12873088682537806 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
6295 |
0.12749542677245987 |
No Hit |
| CATGACTATAAATATGAAGCAAATATGATTATCCTCACAAGTTCTGGAAT |
6276 |
0.12711061134614107 |
No Hit |
| GTATCTTAATGATGTATATAATTGCCTTCAATCCCCTTCTCACCCCACCC |
6251 |
0.12660427525887952 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCACTGCAGCCTGCAGAGCCACA |
6216 |
0.12589540473671332 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACATTTTAGAGATATCTGGGTTTTTTTC |
6201 |
0.1255916030843564 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
6155 |
0.12465994468379513 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
6130 |
0.12415360859653359 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
6084 |
0.12322195019597232 |
No Hit |
| CATGACTATAAATATCAAGCAAATATGATTTTACTCACAAGTTCTGGAAT |
6077 |
0.1230801760915391 |
No Hit |
| TGTGGCTCTGCAGGCTGCAGTGAGACCGCACCAGCGACACA |
6034 |
0.12220927802144921 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6026 |
0.1220472504735255 |
No Hit |
| CATGTCCTCCTTTCTACCAATAACCGCATATCTACTCTTTTTCTGTACTT |
6023 |
0.12198649014305413 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
6019 |
0.12190547636909228 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
6019 |
0.12190547636909228 |
No Hit |
| ATCATGTCCCCAGAAAATCCAGTTACCTCTTGGTCAGTCATCCGGAAGCA |
6012 |
0.12176370226465905 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATCTGGGTTTTTTTC |
5999 |
0.12150040749928304 |
No Hit |
| TAATAGGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGT |
5985 |
0.12121685929041656 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
5970 |
0.12091305763805961 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
5804 |
0.11755098601864289 |
No Hit |
| GAGTTGACTTTCTAAAAGGTGCTATTTCTTTTTCTTTTCTCTGCTTAGGA |
5741 |
0.11627501907874377 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
5725 |
0.11595096398289637 |
No Hit |
| CATGACAGTTGCTTTGTATATTTTCAAGCACGGCTGGAGGTGCTACTTCT |
5709 |
0.11562690888704896 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
5676 |
0.11495854525186372 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
5674 |
0.1149180383648828 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
5607 |
0.11356105765102183 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
5596 |
0.11333826977262675 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
5517 |
0.11173824773688021 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
5424 |
0.10985467749226724 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5382 |
0.10900403286566783 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
5367 |
0.10870023121331089 |
No Hit |
| GACTGAACAAGTAGGAAAATGACAGAGTTATCTTCCCAATTGATGTAAGT |
5340 |
0.10815338823906842 |
No Hit |
| ATGTCAGATATAAAGAACTCACCGAACAGCAGCTCCCAGGCGCACTTCCT |
5319 |
0.1077280659257687 |
No Hit |
| GACTGAAAGAGGGAGAAATAAAGACAAGGCCCCCGAGGAGCTGTCCAAAG |
5313 |
0.10760654526482594 |
No Hit |
| TGATCCCTTTTTCCTTTTCTCTTTCCTTGACTCCCGCTTATTCTCCTTTT |
5301 |
0.10736350394294038 |
No Hit |
| TAAAACATCGCCTACAGAAAAGCGTATGAAAGGAGTGTAAGCTTTGCTCT |
5292 |
0.10718122295152621 |
No Hit |
| CATGGAGGAAGTAGGCAGCTGACAGAAAGTGCTGCAATTGCCTGTGTCAA |
5289 |
0.10712046262105483 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
5286 |
0.10705970229058345 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
5277 |
0.10687742129916929 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
5251 |
0.10635083176841727 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
5180 |
0.10491283728059445 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCAGCTTGGCGCAATCATGACACA |
5147 |
0.10424447364540919 |
No Hit |
| AACTTTATAAACCAACTTCTGCTAGCTTCCAACTTCTCTTCTGCAGCTTC |
5079 |
0.10286723948805777 |
No Hit |
| TAATCCAGAGCAAGGCCTTGACTCCCTTCAGTTCTATGAAGAGAATGAGG |
5064 |
0.10256343783570084 |
No Hit |
| TGTGTCATGATTGCGCCAAGCTGAGACCGCACCAGCGACACA |
5053 |
0.10234064995730573 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
4947 |
0.10019378494731675 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality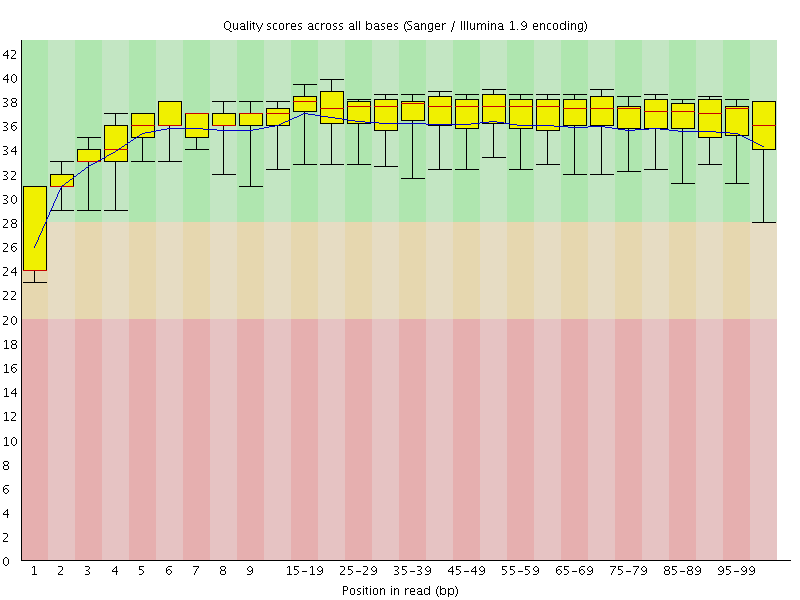
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores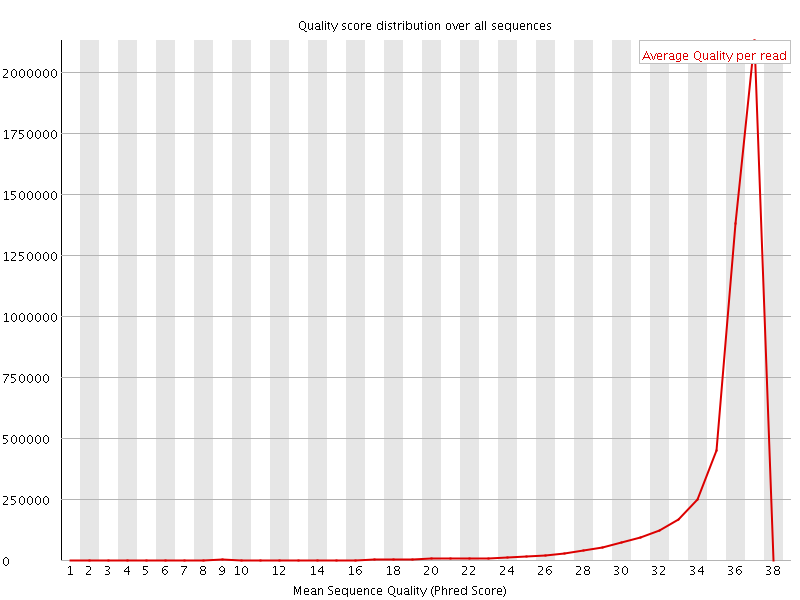
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content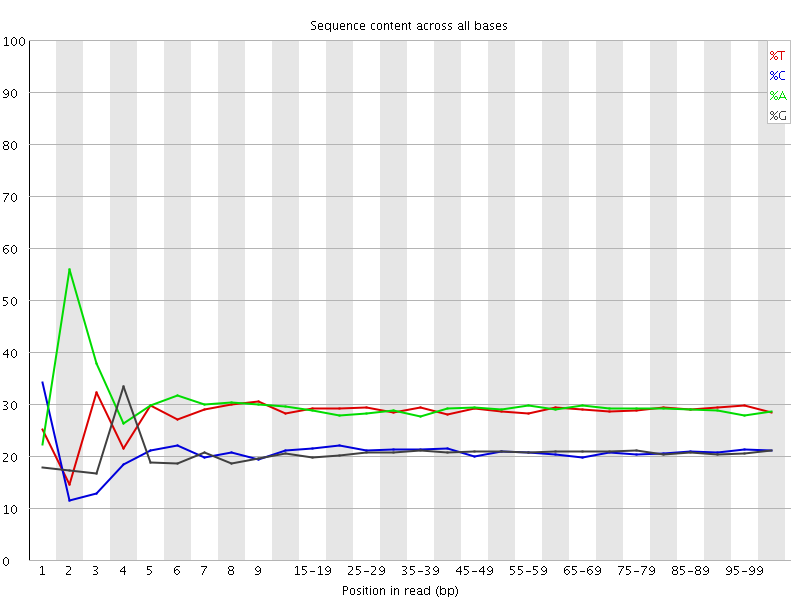
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content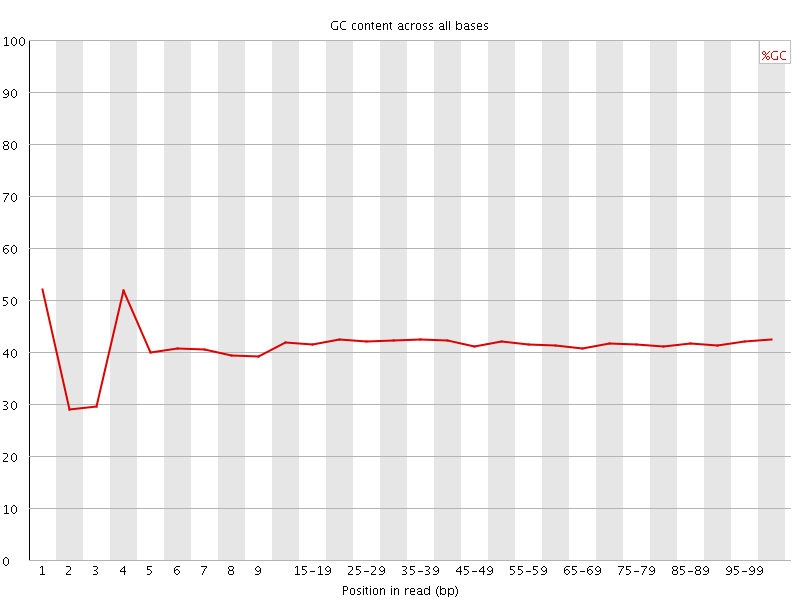
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content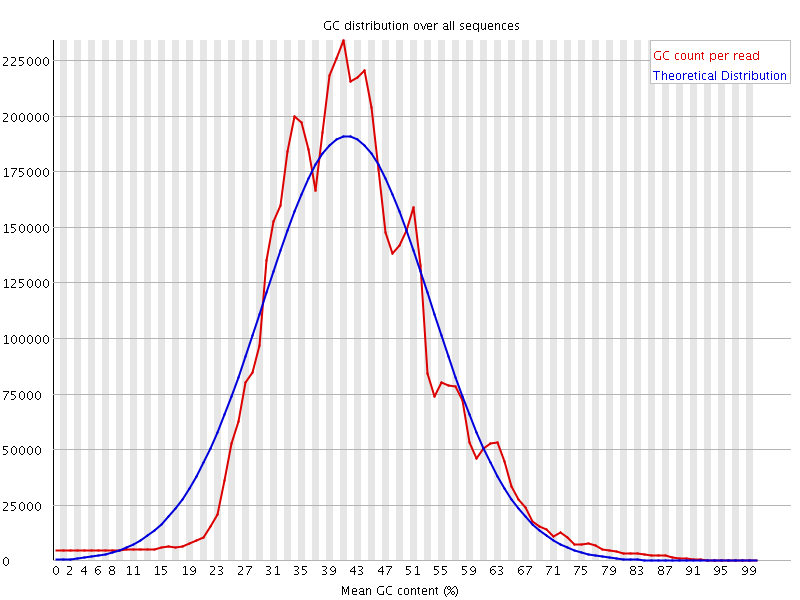
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution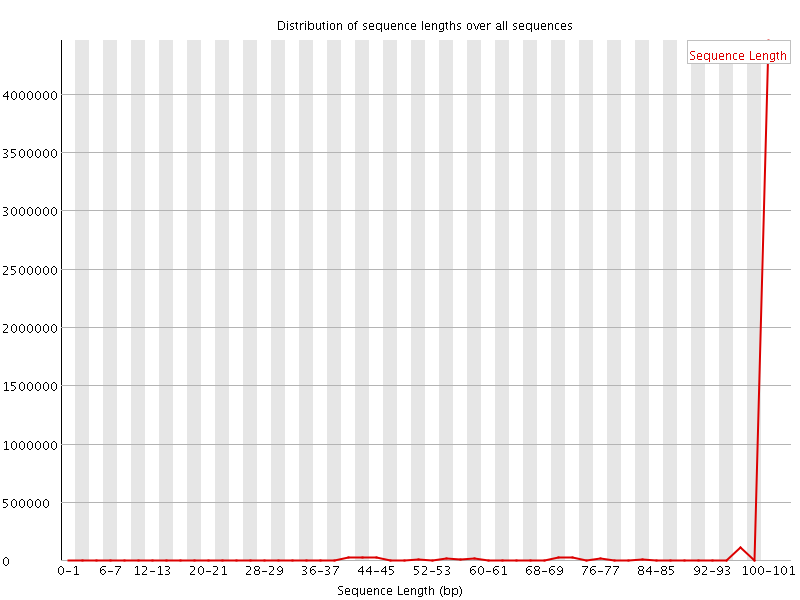
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels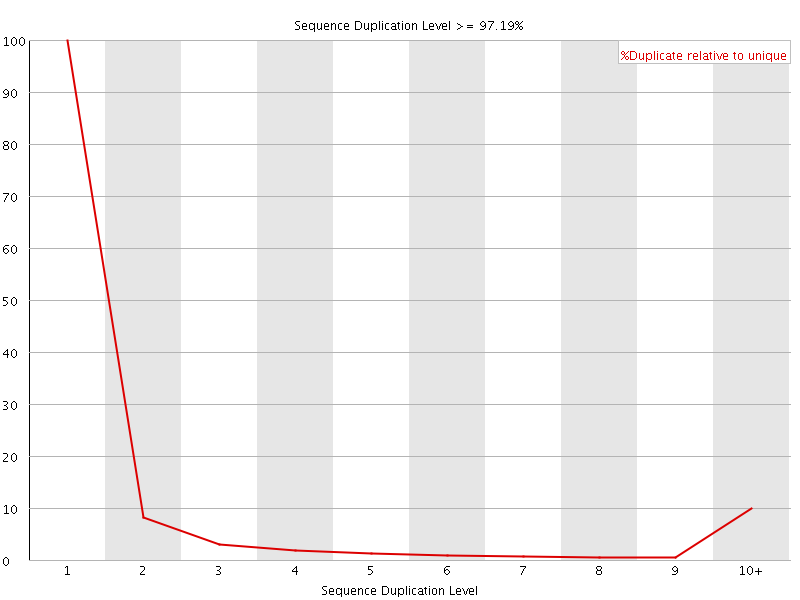
![[WARN]](Icons/warning.png) Overrepresented sequences
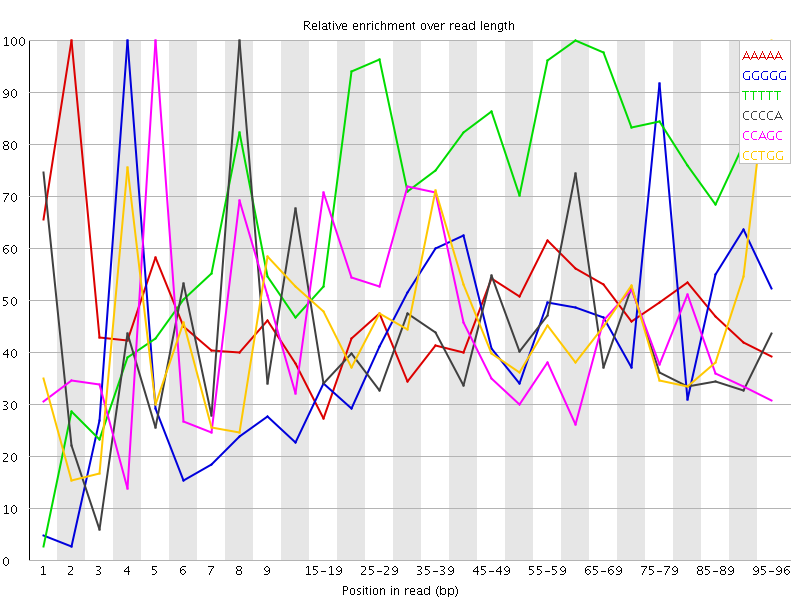
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content