| Sequence |
Count |
Percentage |
Possible Source |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
26051 |
0.2664535120395543 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
25814 |
0.26402944070435125 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
25372 |
0.25950859880494304 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
25165 |
0.2573913719425505 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
19343 |
0.197843087919124 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
19198 |
0.19636000630053985 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
19113 |
0.19549061362757672 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
18867 |
0.19297448895053054 |
No Hit |
| * |
18314 |
0.18731832250172345 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
16618 |
0.16997138163883588 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
16340 |
0.16712795619079182 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
16001 |
0.16366061364803303 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
15873 |
0.1623514105640415 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
15871 |
0.16233095426585414 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
15788 |
0.16148201789107838 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
15771 |
0.16130813935648577 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
15724 |
0.16082741634908262 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
15696 |
0.1605410281744595 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
15408 |
0.1575953212354786 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
15262 |
0.15610201146780078 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
13933 |
0.1425088013222951 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
13817 |
0.1413223360274278 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
13801 |
0.14115868564192885 |
No Hit |
| TGAGACCGCACCAGCGACACA |
13514 |
0.13822320685204165 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
13470 |
0.13777316829191957 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
13387 |
0.1369242319171438 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
13338 |
0.1364230526115533 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
12978 |
0.13274091893782716 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
12870 |
0.13163627883570933 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
12738 |
0.13028616315534308 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
12610 |
0.12897696007135157 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
11846 |
0.12116265416377721 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
11706 |
0.11973071329066148 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
11590 |
0.11854424799579417 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
11491 |
0.1175316612355195 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
11274 |
0.11531215288219014 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
11115 |
0.11368587717629443 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
10738 |
0.10982986496797566 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
10629 |
0.10871499671676414 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
10611 |
0.10853089003307784 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
10571 |
0.10812176406933048 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
10537 |
0.10777400700014524 |
No Hit |
| ATTCTTACTTGTTTGTTTGTTTGTTTGTTTGTTTTTTGTAGGGTACAGGC |
10535 |
0.10775355070195788 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
10509 |
0.10748761882552209 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
10495 |
0.10734442473821053 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
10465 |
0.1070375802654 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
10296 |
0.10530902306856746 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
10291 |
0.10525788232309904 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
10281 |
0.10515560083216222 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
10266 |
0.10500217859575696 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
10073 |
0.10302814582067599 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
10023 |
0.1025167383659918 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
9975 |
0.102025787209495 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
9903 |
0.10128936047474976 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality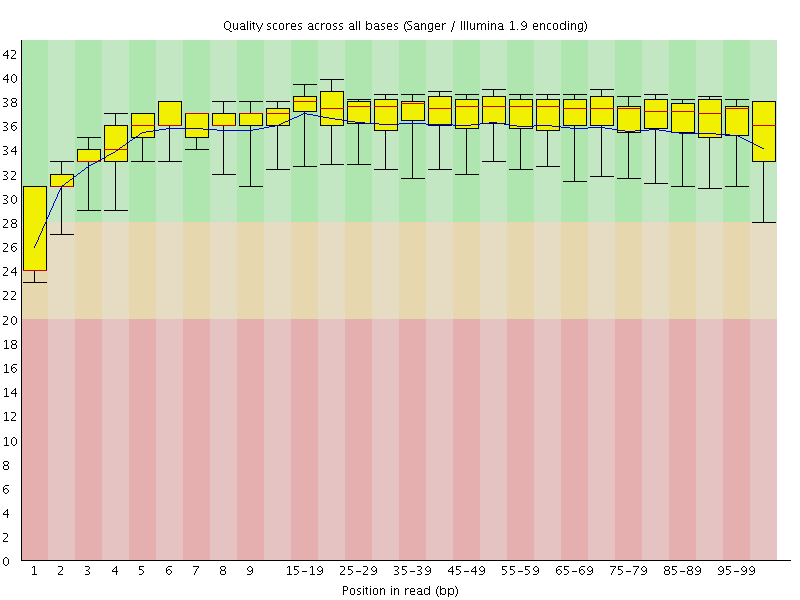
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores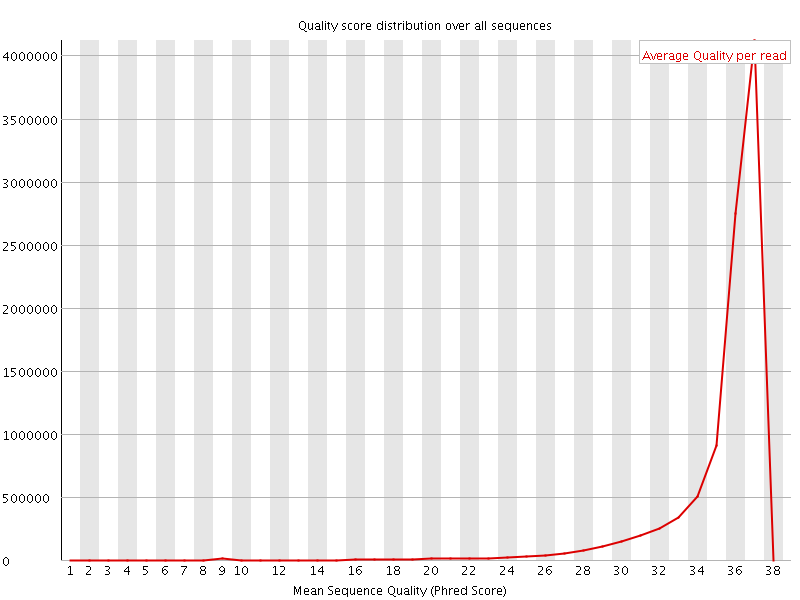
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content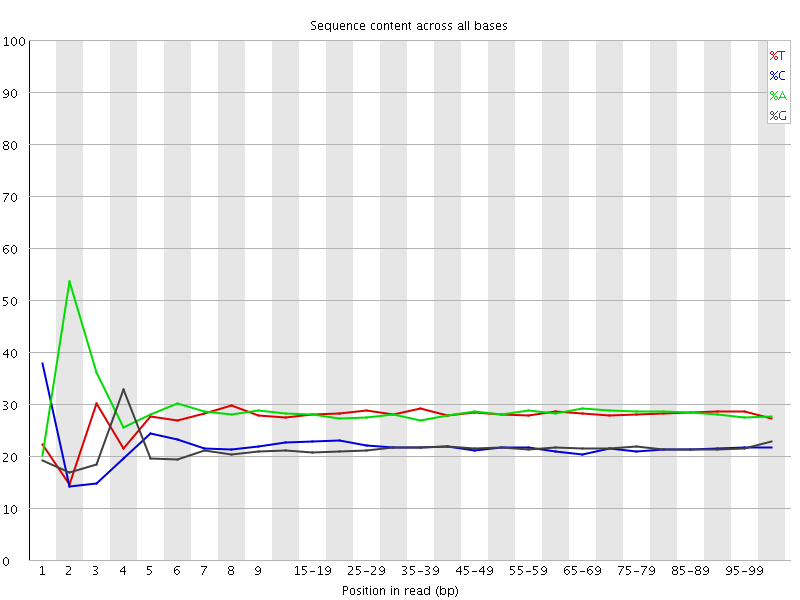
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content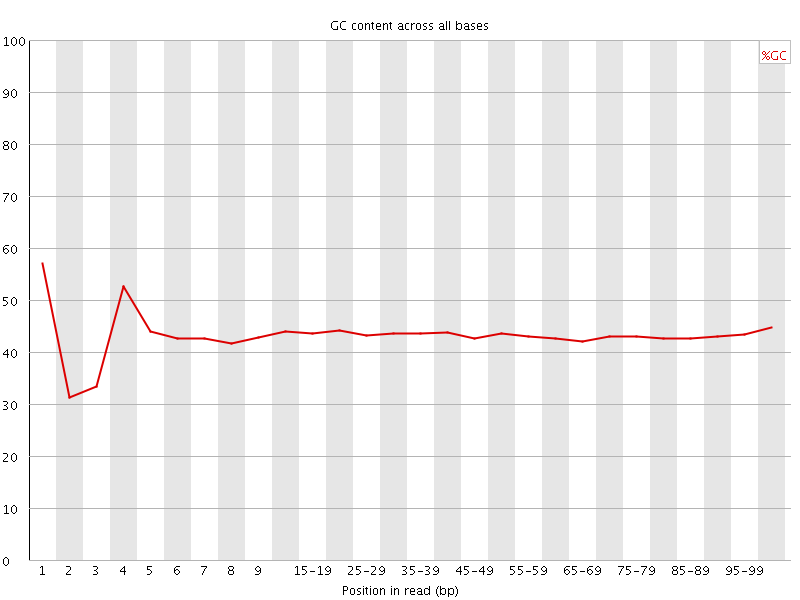
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content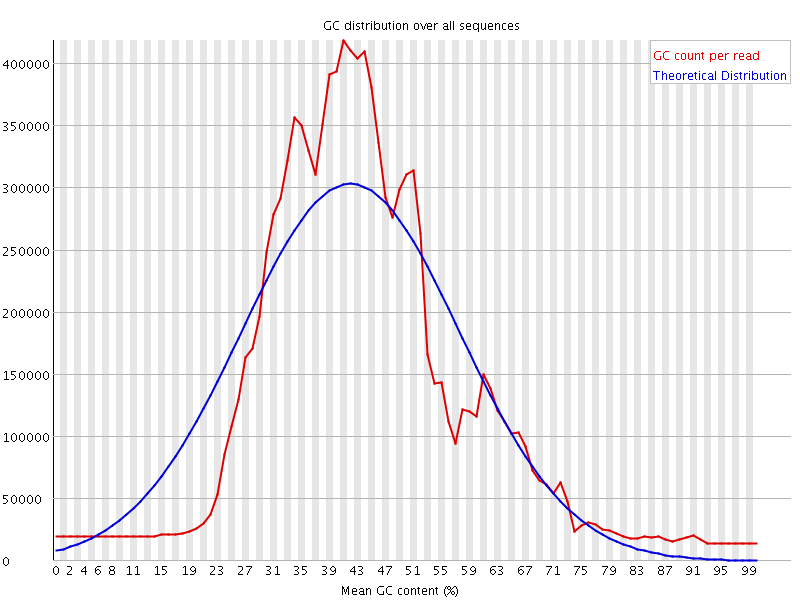
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
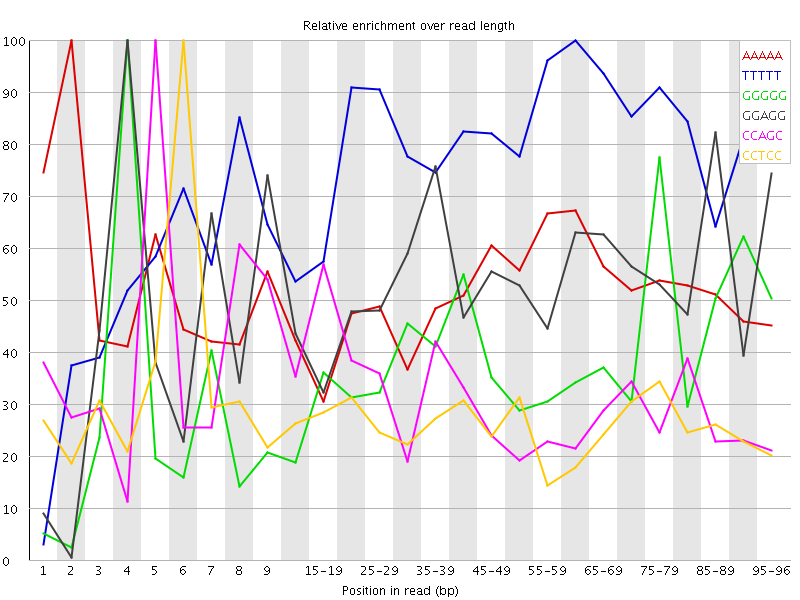
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content