| Sequence |
Count |
Percentage |
Possible Source |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10457 |
0.21901375323431183 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
10360 |
0.2169821634797237 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10316 |
0.21606061761166306 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
10227 |
0.2141965816512678 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
10168 |
0.21296087241909561 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
10096 |
0.21145288827136005 |
No Hit |
| * |
9616 |
0.2013996606197899 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
9065 |
0.18985939304475824 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
8938 |
0.18719947656194694 |
No Hit |
| TGAGACCGCACCAGCGACACA |
8275 |
0.1733134558682156 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
8178 |
0.17128186611362745 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
7602 |
0.1592179929317432 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
7547 |
0.15806606059666745 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7415 |
0.15530142299248564 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
7222 |
0.15125918770758345 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7160 |
0.14996064580258894 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
7047 |
0.14759394845961513 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
7044 |
0.14753111578679282 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
7037 |
0.1473845062168741 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7020 |
0.14702845440421433 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
7013 |
0.1468818448342956 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
6998 |
0.146567681470184 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
6975 |
0.1460859643118796 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
6938 |
0.14531102801373774 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
6934 |
0.1452272511166413 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
6892 |
0.14434759369712893 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
6751 |
0.14139445807448017 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
6702 |
0.14036819108504905 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6682 |
0.13994930659956695 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
6414 |
0.13433625449410694 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
6397 |
0.13398020268144714 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6344 |
0.13287015879491962 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
6170 |
0.1292258637712254 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
6124 |
0.12826242945461658 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6092 |
0.12759221427784523 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6050 |
0.12671255685833285 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
5900 |
0.12357092321721716 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
5833 |
0.12216766019085214 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
5703 |
0.11944491103521855 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
5687 |
0.11910980344683288 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
5671 |
0.11877469585844719 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
5632 |
0.11795787111175712 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
5593 |
0.11714104636506704 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5545 |
0.11613572359991002 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
5533 |
0.11588439290862076 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
5502 |
0.11523512195612354 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
5500 |
0.11519323350757532 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
5486 |
0.11490001436773785 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
5465 |
0.11446018565798165 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
5292 |
0.11083683485856156 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
5116 |
0.10715065138631914 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
5076 |
0.10631288241535497 |
No Hit |
| CATGGAATATATATTCTCCTGGCTGGTGCTTATGATTGCAAAACTTGGAT |
5069 |
0.10616627284543623 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
5040 |
0.10555889034148722 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
4974 |
0.10417657153939629 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
4938 |
0.10342257946552853 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
4928 |
0.10321313722278748 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
4923 |
0.10310841610141697 |
No Hit |
| TGACTGGCAATTGTGTCAACAGGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTC |
4911 |
0.1028570854101277 |
No Hit |
| GACAACTTGGCCACCGACAGCTGAAGGGCTTTTCACCTGTTGACACAATTGCCAGTCA |
4909 |
0.1028151969615795 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
4906 |
0.10275236428875717 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
4887 |
0.1023544240275492 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
4868 |
0.10195648376634121 |
No Hit |
| CAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCTCTAGACAAC |
4810 |
0.10074171875844314 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
4781 |
0.10013433625449411 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality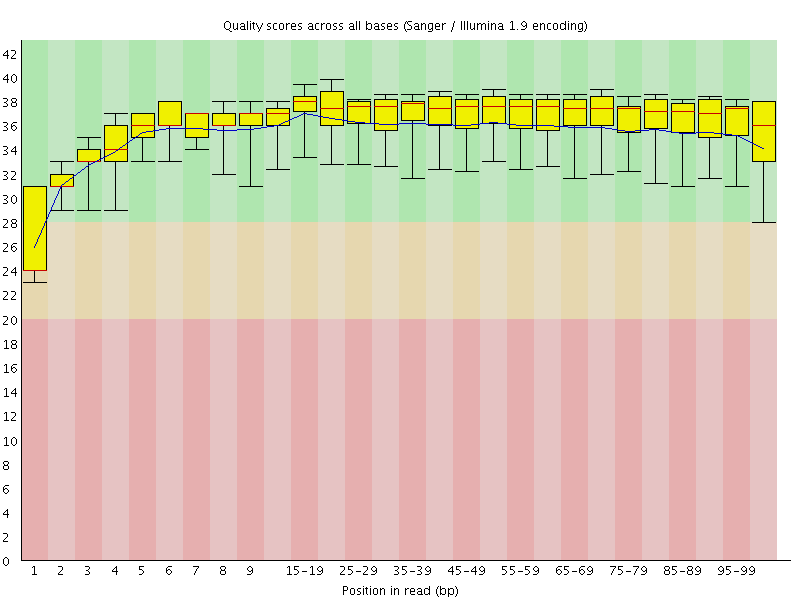
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores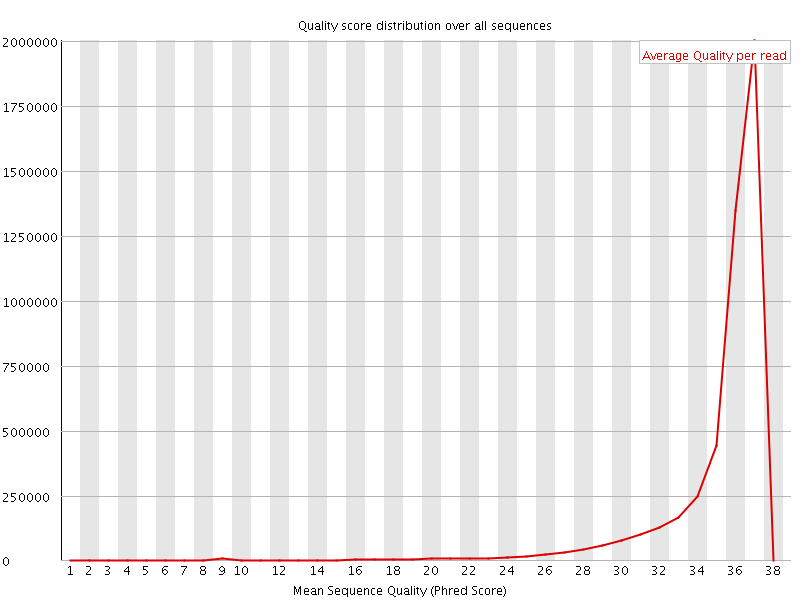
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content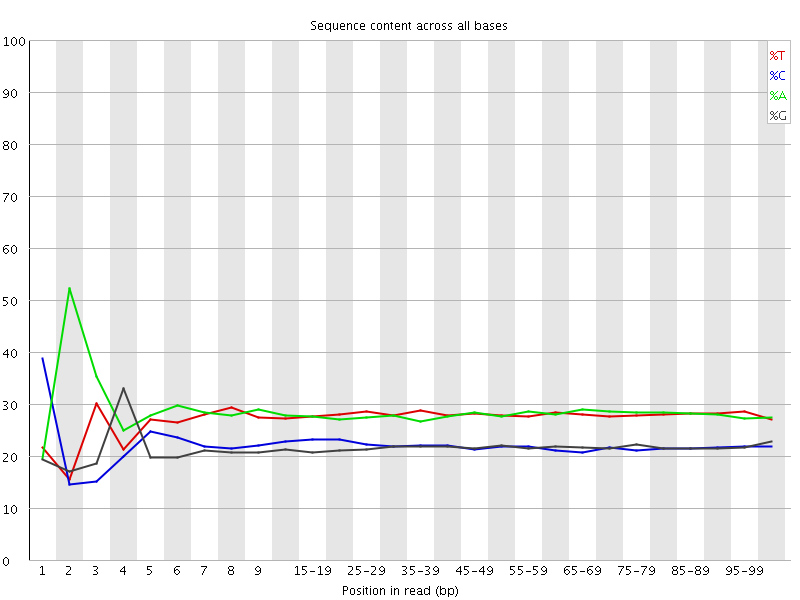
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content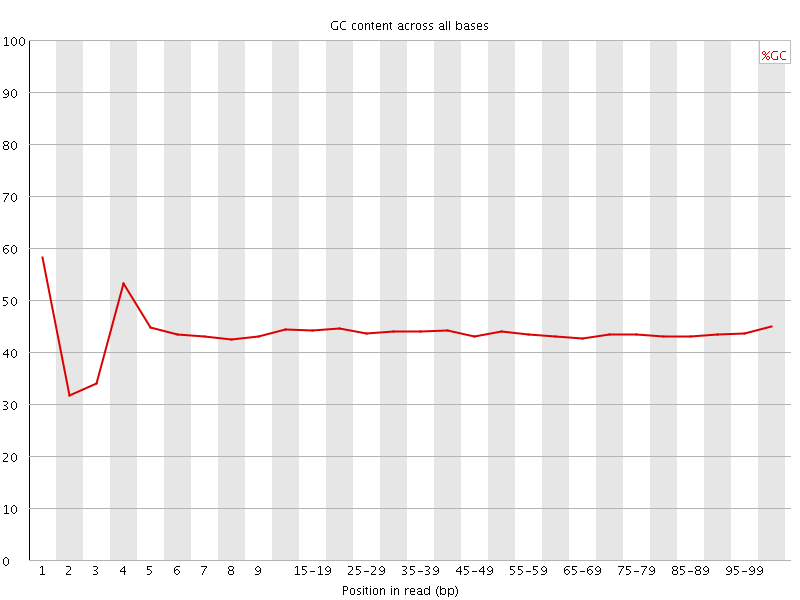
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content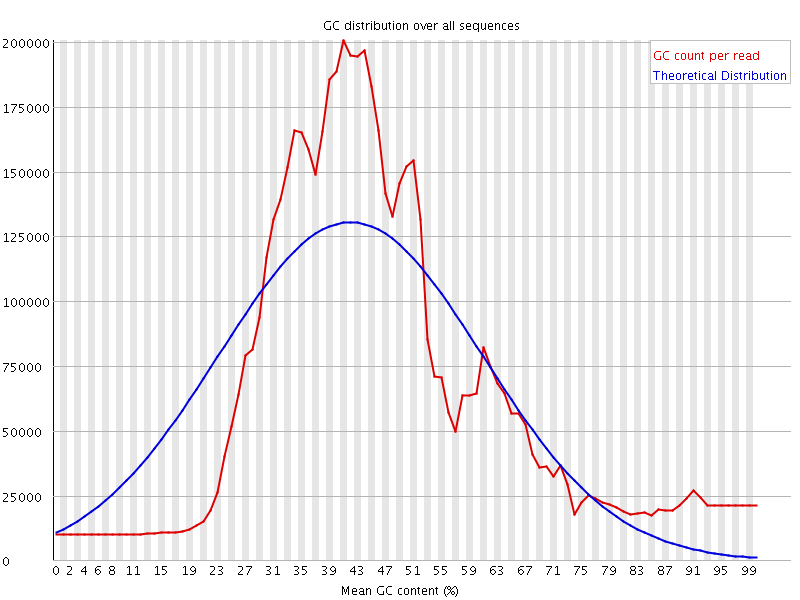
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution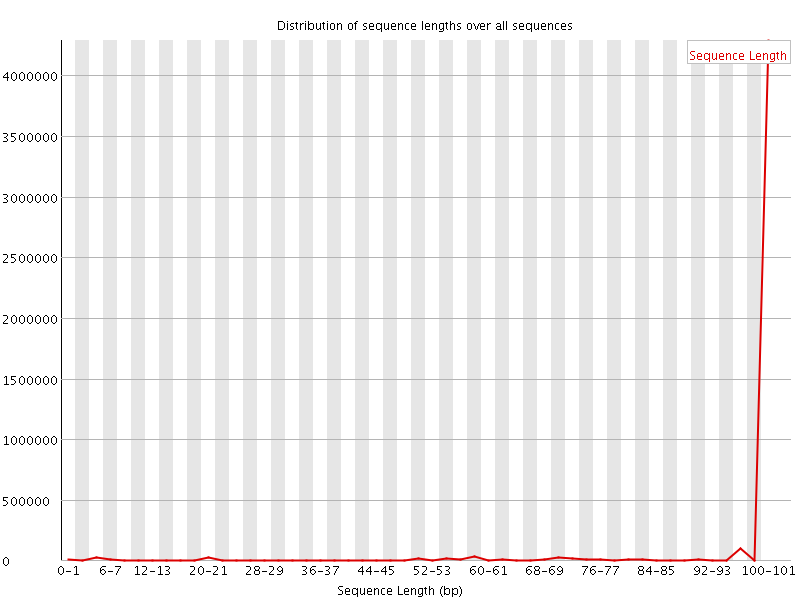
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels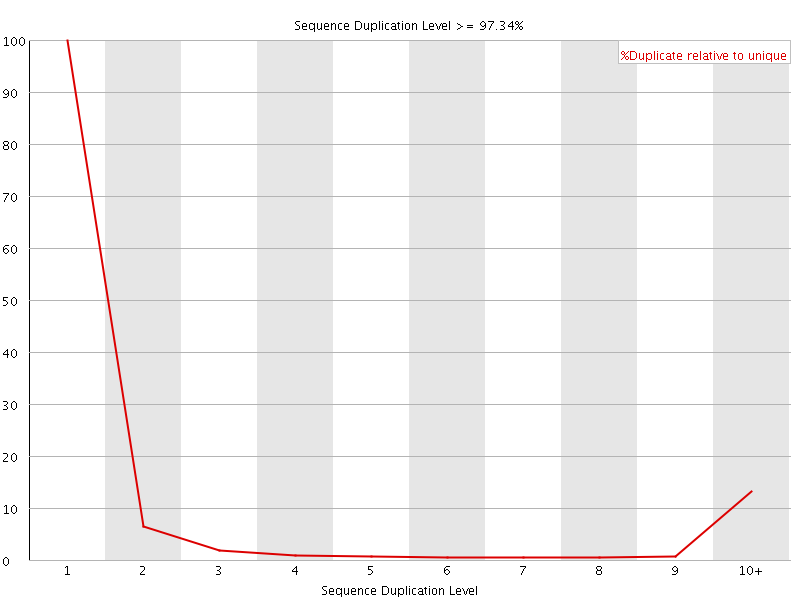
![[WARN]](Icons/warning.png) Overrepresented sequences
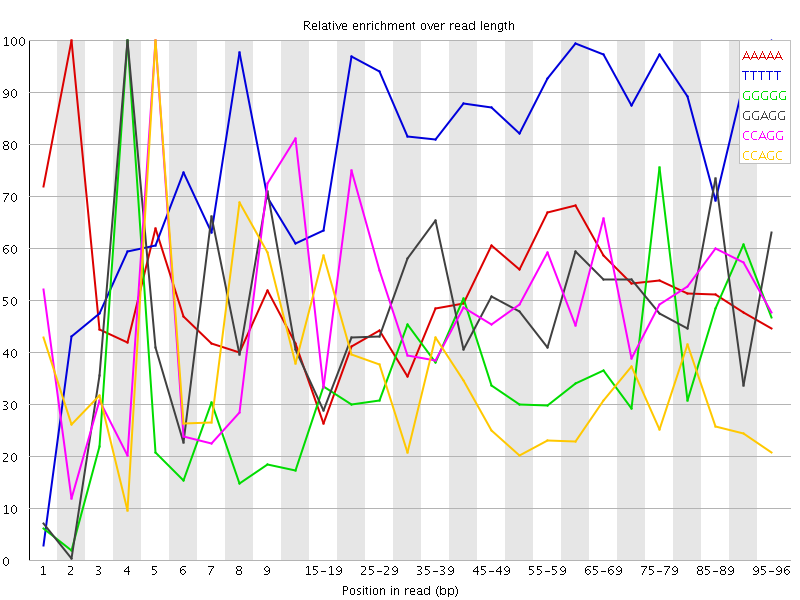
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content