| Sequence |
Count |
Percentage |
Possible Source |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
16955 |
0.28893055987261496 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
16807 |
0.28640848833848653 |
No Hit |
| TGAGACCGCACCAGCGACACA |
12231 |
0.2084287630670571 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCA |
12040 |
0.20517392750612115 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
11977 |
0.20410034300172866 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
11890 |
0.20261777392423425 |
No Hit |
| * |
11329 |
0.19305775952797727 |
No Hit |
| CACTCAGTATGAGGAGCCGGCTGTCATCACATAGGGACAGGGCTCTCCCT |
11028 |
0.18792841134032426 |
No Hit |
| CATGCCCTCCAGCGGCCCCGGGGACACCAGCAGCTCTGCTGCGGAGCGGG |
11015 |
0.18770687802989405 |
No Hit |
| CATGCCAGCTACGATTACGAAACCAATCAGCAAAGGAGTGAACAGGGTGT |
10703 |
0.18239007857956932 |
No Hit |
| GGCTGTGAGTTGGGAGGTGGGGTCAGTTTGGGACTGAGTGGCTGTGGTAG |
10576 |
0.1802258685469051 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10550 |
0.17978280192604468 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10504 |
0.1789989148275994 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
10242 |
0.17453416657123694 |
No Hit |
| CATGGGCTCGGCCACGCGCTACCACACCTACCTGCCGCCGCCCTACCCCG |
10215 |
0.1740740589264973 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
10167 |
0.17325608978029347 |
No Hit |
| GAGAACTGGTAGGAGCCGGCCGAGGCGCCGTAGTACAGGTGGTAGGAGGG |
10049 |
0.17124524896254245 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
9326 |
0.15892458869784765 |
No Hit |
| CATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTAAGCTGTCCCACT |
9233 |
0.15733977347707775 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
9210 |
0.1569478299278551 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
9208 |
0.15691374788009663 |
No Hit |
| TTTGCCCAAGACTGGACAGCGGGCACACTTACCAGTGTGCTTCCTGCTGT |
9126 |
0.15551638392199846 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
9055 |
0.15430647122657198 |
No Hit |
| GACGCGACCGGACCGAGCGCCAAACGCGGCAGCCCAATCGCCATCGCTTT |
8766 |
0.14938161532546992 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
8661 |
0.1475923078181491 |
No Hit |
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG |
8594 |
0.14645055921823963 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
8570 |
0.1460415746451377 |
No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA |
8561 |
0.14588820543022452 |
No Hit |
| CGAGCCCGGCGGGCCCTGTGATTGGACGGGCGCCCGCCTCGCGTCCCGCC |
8330 |
0.1419517289141187 |
No Hit |
| GAGGGAGAGGCCGGAGCTGCCCAGGACAGGCTCTGGGTCCTGCCCTTGCA |
8065 |
0.1374358575861185 |
No Hit |
| CATGGCTCCACTCTCCACCGTCTCCACGCACACCTTCTCCAGCATCTGGG |
8033 |
0.13689054482198265 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
7833 |
0.13348234004613346 |
No Hit |
| TGAGTAGGGGGAGCAAATCGTGCCTTGTCATTTTATTTGGAGGTTTCCTG |
7800 |
0.13291998625811835 |
No Hit |
| CCACCGAGCAAACGCATCTTGCTTTTCACATCTCTCCTCCTACAGCCTTA |
7754 |
0.13213609915967303 |
No Hit |
| CATGGAGGGGAGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTC |
7720 |
0.13155670434777866 |
No Hit |
| GCCTCAGCCTTCCTTCGCAGAAAGTTGAGCAGGTCGCCATAGCAACAGTA |
7713 |
0.13143741718062396 |
No Hit |
| GCCCTCCTCCTGCCACGGGCCTGCTCCCCTCCTTCTCTCATGGGGGTCTG |
7699 |
0.1311988428463145 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
7688 |
0.1310113915836428 |
No Hit |
| CATGAATCCAGAAGCACTTAACACTGGTGGTGGTGATTTGTCACATATGG |
7661 |
0.13055128393890317 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
7626 |
0.12995484810312954 |
No Hit |
| AGCACTCAGCACCTCCCTCACCCCACACCCTTGGCTGCTCTAGGCCCTGT |
7521 |
0.12816554059580873 |
No Hit |
| CATGGTAAAGGGTTTGTGCTAACTGACCTCTTCTTTCCTTGCAGGGCTAA |
7510 |
0.127978089333137 |
No Hit |
| CAGGCTGGGTCCCAGCATGGCCTCAGCCTTCCTTCGCAGAAAGTTGAGCA |
7490 |
0.1276372688555521 |
No Hit |
| GTGCTAATGGACCAGGGTCACAGTTTCAAAACTTGAACAATCCAGTTAGC |
7460 |
0.12712603813917472 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
7142 |
0.12170699254557452 |
No Hit |
| CAGCTGGGTGGCCCCAGGAGAGGCGAGGCCCTGAGAGAAAGGCTTTCTAC |
6996 |
0.11921900305920462 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6978 |
0.11891226462937819 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6532 |
0.11131196797923448 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
6459 |
0.11006797323604954 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
6441 |
0.10976123480622312 |
No Hit |
| CCGCGGCGCAGATCGAAACAGATCGGGCGGCTCGGGTTACACACGCACGC |
6342 |
0.10807417344217776 |
No Hit |
| TGCTCTGTGGTCCGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTG |
6324 |
0.10776743501235134 |
No Hit |
| CGAGGCGAGCCGTGAAGTTGCGTGTGCGTGGCAGTGTGCGTGGCAGGATG |
6316 |
0.10763110682131738 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
6231 |
0.10618261979158147 |
No Hit |
| CATGTACAAGGCCTTGCTACCTCAGCAGTCCTACAGCTTGGCCCAGCCGC |
6146 |
0.10473413276184557 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6139 |
0.10461484559469082 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
6138 |
0.10459780457081158 |
No Hit |
| CATGGGTATTATTTCTTTGCTTTTTTTGTGTGGTGGGGGGCTTTATTTGC |
6136 |
0.1045637225230531 |
No Hit |
| GAGGCACGAACACCCACGAGCGCCGCGTAACCGCAGCAGGTGGAGCGGGC |
6120 |
0.10429106614098516 |
No Hit |
| GAAGCAGTAGTTTCACTTCTAGCTGGTCTCCCTCTGCAGCCTGAAGAAGG |
6092 |
0.10381391747236628 |
No Hit |
| GGTGATGCCAAGGACGATGGGATGTGGGGACCGACGTAGTGAGGTGGCGG |
6049 |
0.1030811534455587 |
No Hit |
| AGGGTCAGCCCACACCACCCAGCCCCCACGCCCAACCCTCGTGCCTCCCT |
6007 |
0.10236543044263037 |
No Hit |
| CATGGGGGCCAGTTGTGGGTGGTGGCCCAGGTGCAGGAGAGGCGGGCAGT |
5983 |
0.10195644586952846 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
5960 |
0.10156450232030581 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
5911 |
0.10072949215022275 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality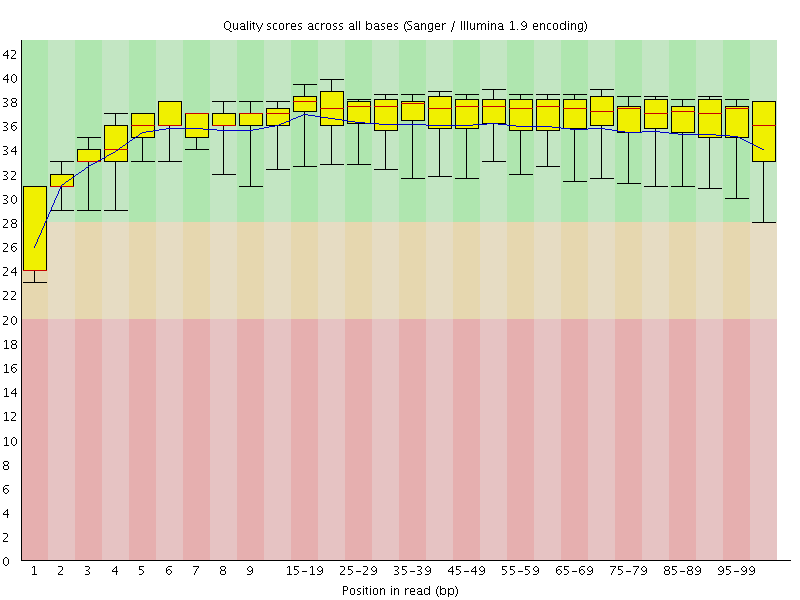
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores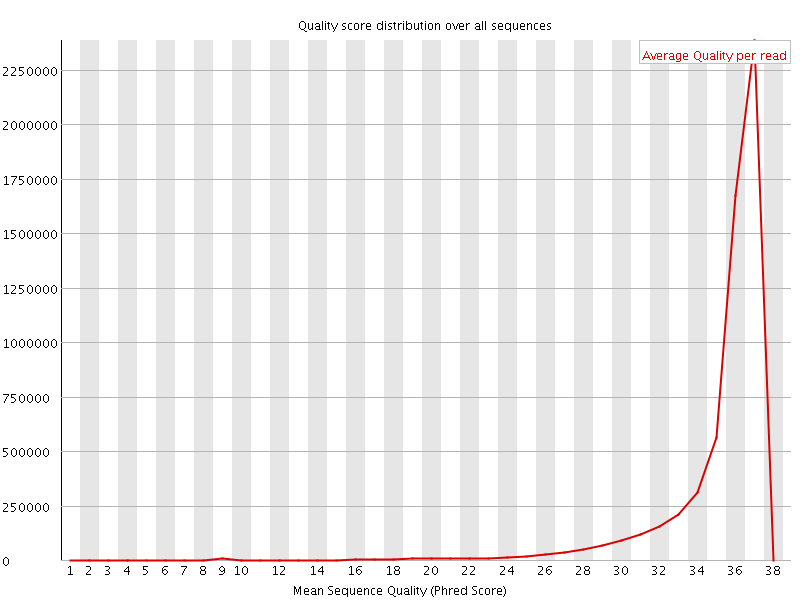
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content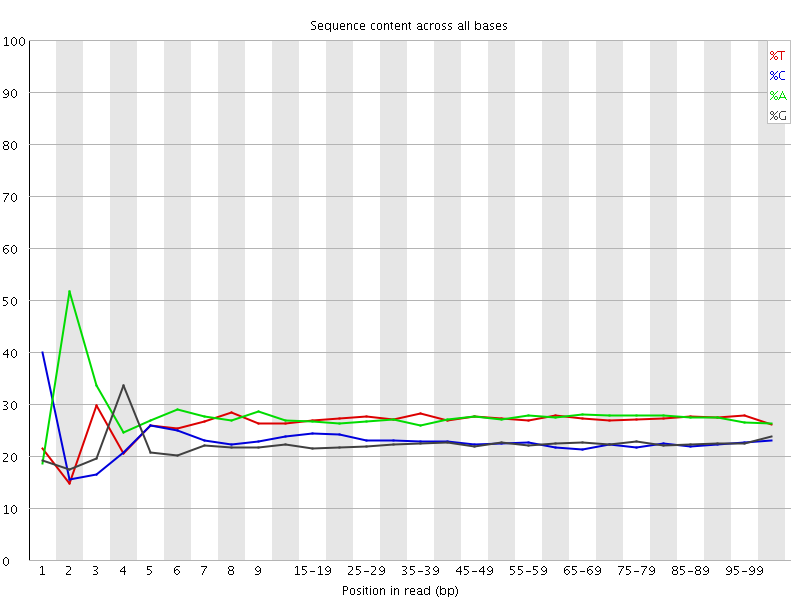
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content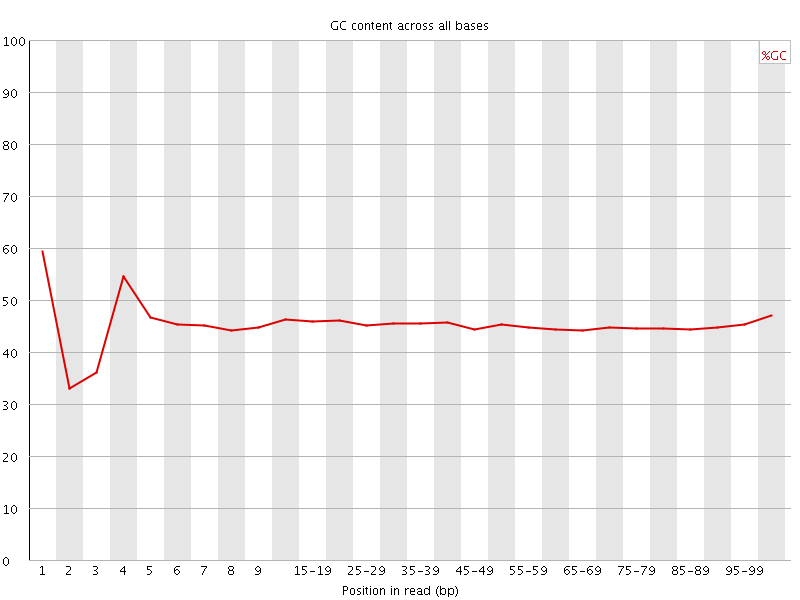
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content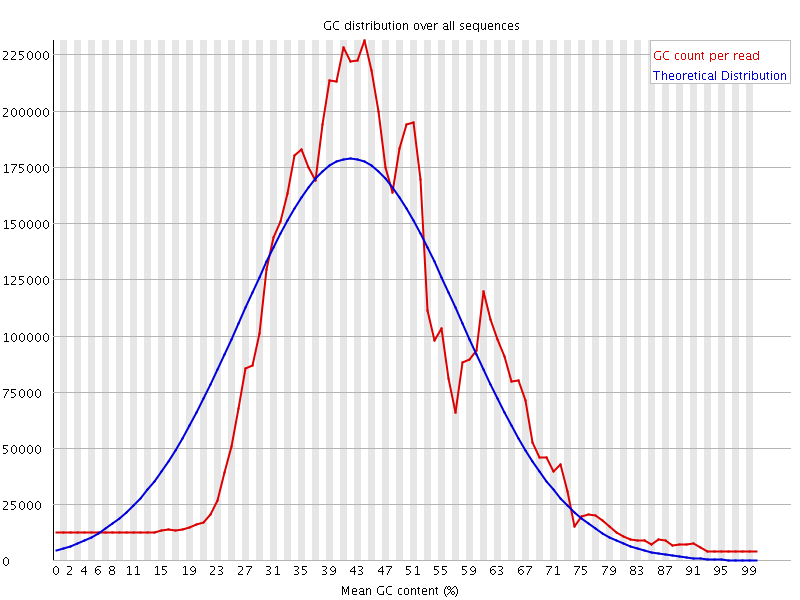
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution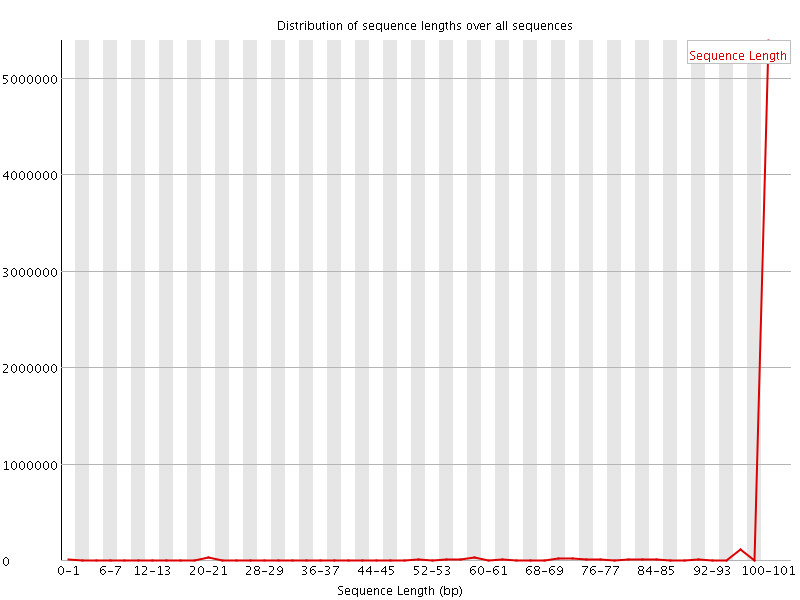
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels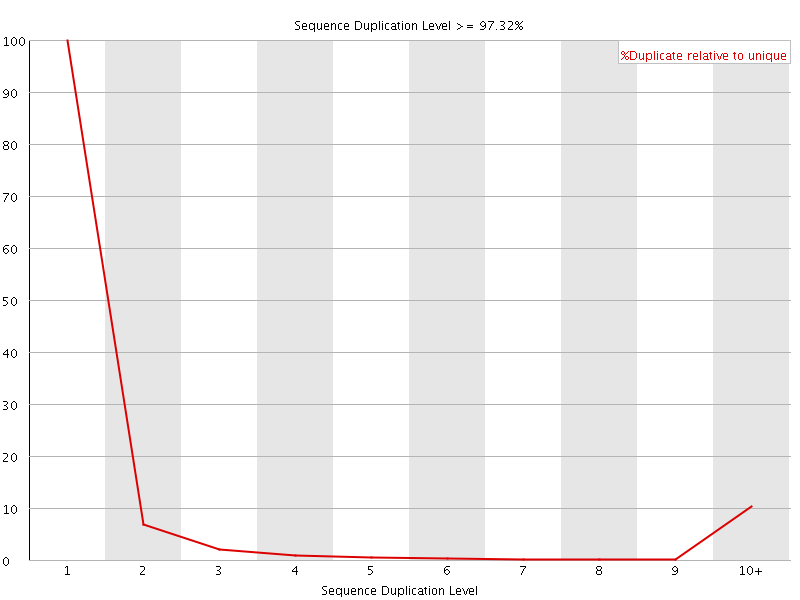
![[WARN]](Icons/warning.png) Overrepresented sequences
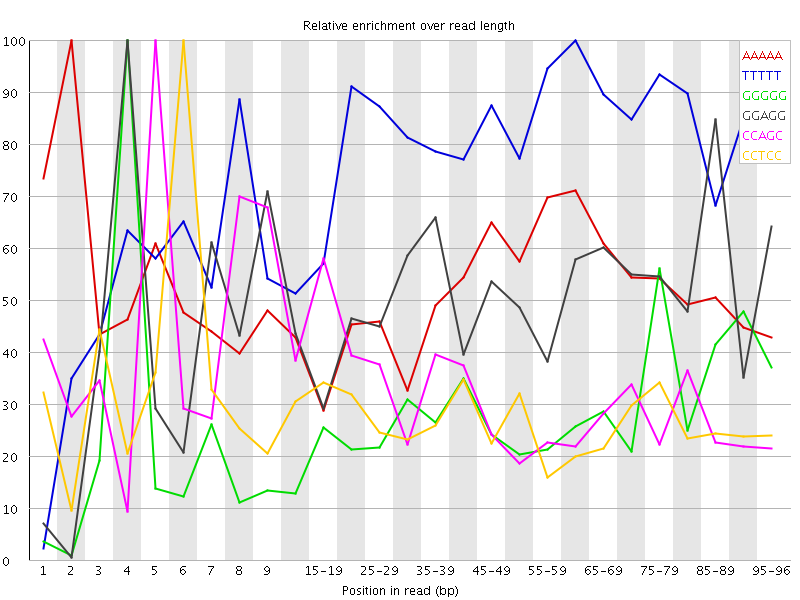
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content