| Sequence |
Count |
Percentage |
Possible Source |
| TGTGTCGCTGGTGCGGTCTCACTGCAGCCTGCAGAGCCACA |
15964 |
0.35958907195205236 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCACGAGACTGCCCTTAACCTACACA |
15846 |
0.35693112215937245 |
No Hit |
| TGTGTAGGTTAAGGGCAGTCTCGTGAGACCGCACCAGCGACACA |
15551 |
0.3502862476776727 |
No Hit |
| TGTGGCTCTGCAGGCTGCAGTGAGACCGCACCAGCGACACA |
15546 |
0.35017362268645746 |
No Hit |
| TGTGTCGCTGGTGCGGTCTCAGCTTGGCGCAATCATGACACA |
13834 |
0.3116108256943556 |
No Hit |
| TGTGTCATGATTGCGCCAAGCTGAGACCGCACCAGCGACACA |
13589 |
0.3060922011248083 |
No Hit |
| AGATTGTTGAGGGATTGCGAGCTGCAGCCAGAAGGAATGGAGATGTTATA |
10732 |
0.24173828114441406 |
No Hit |
| GAGGAAAGGCAGATAGTAATCCTTAAAGGCTGAGGATTGGAACCCAGATC |
10630 |
0.23944073132362292 |
No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA |
9083 |
0.2045945590416244 |
No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA |
8942 |
0.20141853428935433 |
No Hit |
| AAAAATAATAAACCGTTCATTTCTCAGGATGTGGTCATAGAATAAAGTTA |
8718 |
0.1963729346829111 |
No Hit |
| TAAAGAAACCCACAGAAACACACACACATACAATTAGCAAATGAATTCAA |
8629 |
0.19436820983927963 |
No Hit |
| AGCAACAAGAACAATACAAACCTCATTTCCTTTAGTCCTTCTGCCTCTAT |
8537 |
0.19229591000091903 |
No Hit |
| TAACTGTGGAAAAACTAAGTTGTCACCAGCGGTACCTCATAGCATAACTT |
8501 |
0.1914850100641692 |
No Hit |
| TTAGCTTTGAAGTGGATCCTACCAGTTTTATCATCTTTTCACATAAAAGT |
8045 |
0.18121361086533835 |
No Hit |
| GATACAACATTCAAGACAGAGGAACTGGATGCCAAGCCAGACACAGACCC |
7841 |
0.17661851122375613 |
No Hit |
| CATGTATTTTGAACCGGCACCCCTGTTACCACAGAGTGTGGGAGGAACTG |
7777 |
0.17517691133620092 |
No Hit |
| CATG |
7733 |
0.1741858114135067 |
No Hit |
| CATGTAGGTTATATATGCTAGAATTGCATTTAATCACTGTGAAAAGACTG |
7223 |
0.16269806230955114 |
No Hit |
| ATAATCCATACCACCAGTATAGCAGCAATTCTAACAGCCCCCCTACTGTC |
7132 |
0.16064828746943358 |
No Hit |
| CATGACTATAAATATAAAGCAAATATGATTATGCTCACAAGCTCTGGAAT |
7036 |
0.15848588763810076 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATGTGGGGTTTTTTT |
6895 |
0.1553098628858307 |
No Hit |
| ACCTATGTTACACCATCTTCAGTGCCAGTCTTGGGCAAAATTGTGCAAGA |
6822 |
0.15366553801408805 |
No Hit |
| * |
6744 |
0.15190858815113012 |
No Hit |
| CATGTAATTCAGGCAGTTATGGGAGCCCTAGAGGGCCTGAGAGTTGCTATTGGAC |
6738 |
0.15177343816167183 |
No Hit |
| TAAAACTGCATCAAGTCATGGGGCATGTGGAAGGTAGGGAGGCAAGATGA |
6703 |
0.15098506322316507 |
No Hit |
| GTCCAATAGCAACTCTCAGGCCCTCTAGGGCTCCCATAACTGCCTGAATTACATG |
6693 |
0.15075981324073456 |
No Hit |
| ATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATGTGGATCTA |
6575 |
0.14810186344805465 |
No Hit |
| ATCCTTCACAAAGTCTTTGAGATATATTTTTATCAAATATTTAGCATGGATCCCGGTACACTTTCAATACTTA |
6493 |
0.14625481359212453 |
No Hit |
| TAAAGTGTTGGTTGTTGTGAGGGCTTATACGAAAGCAAGAAACAAGGCAG |
6490 |
0.14618723859739538 |
No Hit |
| ACAGATTTTGGGGTTGTGTTGTCACCCAAGAGATTGTTGTTTGCCATACT |
6486 |
0.1460971386044232 |
No Hit |
| TAAGTATTGAAAGTGTACCGGGATCCATGCTAAATATTTGATAAAAATATATCTCAAAGACTTTGTGAAGGAT |
6424 |
0.1447005887133541 |
No Hit |
| AGCAATTCCATTTATGTGTTTGTTAGAGGTAAATGCTTGGCTTTCTGCAGTGCTGTGCTTTCAAGAATTTA |
6404 |
0.14425008874849307 |
No Hit |
| TAAATTCTTGAAAGCACAGCACTGCAGAAAGCCAAGCATTTACCTCTAACAAACACATAAATGGAATTGCT |
6348 |
0.14298868884688226 |
No Hit |
| ATTGTTTCAAAAAAAATCAAACTGTAGTTGTTTTGGCGATAGGTCTCACG |
6279 |
0.1414344639681118 |
No Hit |
| CACCAAAAACCCCAAGACAGAAATCTTAGGTATTCAGTTTCTTTTTCACA |
6268 |
0.14118668898743827 |
No Hit |
| CATGATATACATACTCTCTGTAAAGTTACTCTTGGTTGCTTGATAGGTAG |
6038 |
0.1360059393915367 |
No Hit |
| TAATCTACCTGAAGTCAATGAATGCAATTTTTCACACACACACACACACA |
6014 |
0.1354653394337035 |
No Hit |
| AAGATTGATAGACCTGAAGATGCTGGGGAGAAAGAACATGTCACTAAGAG |
6000 |
0.13514998945830084 |
No Hit |
| AAACCTGGAGTTTCTTATGTTGTCCCAACCAAGGCCGACAAAAGGAGATC |
5964 |
0.13433908952155102 |
No Hit |
| TAACTCTGCTTACAGAATGGCAGTTATCCATCTTTGGCTCATTTTTGTGG |
5887 |
0.13260466465683615 |
No Hit |
| TAACTAACTCATAAATGCCTATTTTTTCTTACAGAAAAGGGATTTATAAA |
5860 |
0.1319964897042738 |
No Hit |
| AACCTGACAGAAAAACAAAACTCTTCATATTGGTCAATTTCATAATCTTC |
5855 |
0.13188386471305855 |
No Hit |
| TAAACATTCCACTTCCTCCATAGGCTCCATGTTGGCTCAGGCAGACAAGC |
5787 |
0.13035216483253115 |
No Hit |
| AGACTCTTACTCTTCTCAATCTTGCAGAGCTGAGCTTTGGCCTTTTTTAG |
5762 |
0.12978903987645488 |
No Hit |
| AGGACAACAGCTGATCCATACATTTGCTGACAGGTGTACCTGCGTTTCCC |
5740 |
0.1292934899151078 |
No Hit |
| GACTGAACAAGTAGGAAAATGACAGAGTTATCTTCCCAATTGATGTAAGT |
5729 |
0.12904571493443423 |
No Hit |
| CATGACTATAAATATCAAGCAAATATGATTTTACTCACAAGTTCTGGAAT |
5706 |
0.1285276399748441 |
No Hit |
| CATGGAGGAAGTAGGCAGCTGACAGAAAGTGCTGCAATTGCCTGTGTCAA |
5705 |
0.12850511497660103 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACACTTTAGAGATATCTGGGTTTTTTTC |
5664 |
0.12758159004863598 |
No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA |
5623 |
0.12665806512067093 |
No Hit |
| ATCATGTCCCCAGAAAATCCAGTTACCTCTTGGTCAGTCATCCGGAAGCA |
5557 |
0.1251714152366296 |
No Hit |
| TAATAGGGTGGTTCTCTTCCCAAAGTGGAAGCCAAATTCATCAATTATGT |
5520 |
0.12433799030163675 |
No Hit |
| CAATTCAAATAAAAACGAAACTGTGTCAATTAGTTGAAGTAATGATGGCA |
5348 |
0.12046369060383212 |
No Hit |
| ACTTGAAAGCAAGGATAGAGAAACATTTTAGAGATATCTGGGTTTTTTTC |
5195 |
0.11701736587264547 |
No Hit |
| CATGACTATAAATATGAAGCAAATATGATTATCCTCACAAGTTCTGGAAT |
5187 |
0.11683716588670105 |
No Hit |
| CATGGGGTTGTGCCAATTAATTACCAAACATTGAGCCTGCAGGCTTTGAG |
5163 |
0.11629656592886785 |
No Hit |
| TAAACTTATACAGCGAGAAAATGTCATTGAATATAAACACTGTTTGATTA |
5151 |
0.11602626594995125 |
No Hit |
| GTTTTTATACACTAAGGACACTCCCAGTGATGAAAATGAATTCCCCTCCA |
5124 |
0.11541809099738891 |
No Hit |
| CATGGTCACTCTCCCCAAAATATTATATTTTTTCTATAAAAAGAAAAAAA |
5101 |
0.11490001603779874 |
No Hit |
| TAAATATAGCCCCAAAATGGTTGCTATAATAATCCCCATTTCATACTGGG |
5095 |
0.11476486604834045 |
No Hit |
| AGACTGCTGAAGTTCTTCTTTGTCCCACTGAGGTTGTACTCTTCATTCTC |
5087 |
0.11458466606239605 |
No Hit |
| AAGACAGGGGAAAGGAAGAAAAGGGAGTGAAGAGGGAATGGGAGAGGGAA |
5055 |
0.11386386611861843 |
No Hit |
| CATGTCCTCCTTTCTACCAATAACCGCATATCTACTCTTTTTCTGTACTT |
5044 |
0.1136160911379449 |
No Hit |
| GTATCTTAATGATGTATATAATTGCCTTCAATCCCCTTCTCACCCCACCC |
5021 |
0.11309801617835474 |
No Hit |
| CATGTTGTCAGATTACCCTTTAGAATGCCTGTTGCTTTTAAAATATTTTT |
5020 |
0.11307549118011169 |
No Hit |
| CATGCTTTACTCAACTGAAATAATCAAGCCCCTATTGCTGTGGGGGAAGC |
4990 |
0.11239974123282018 |
No Hit |
| CAGCATAATAAAAAATAGGATTCCCAGCTTTGGAAGCCCCAGCTTGGTAG |
4987 |
0.11233216623809103 |
No Hit |
| TAGACATCTATAACTAACAACCACTTTTCTTACTATCATTGAAGTCAATA |
4830 |
0.10879574151393216 |
No Hit |
| GATGGCTCCAGCAGCATCCTCAGCACCAACAACGCTACCTTCCAAAACAC |
4779 |
0.1076469666035366 |
No Hit |
| AGACTCCTCACCTTTGACATAGAGGTGGATGGCGGCGCTGCCTCCCAGGG |
4740 |
0.10676849167205765 |
No Hit |
| TGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGT |
4675 |
0.10530436678625939 |
No Hit |
| AAGTCCTCCTTATTCAAGATTTTGAAATTCTTAGCCTGGGAGTGCTGGAG |
4623 |
0.10413306687762078 |
No Hit |
| TAACACTGATATACAAATTGGGACTCTTCATTCTGGAGAAAGCATCAGGT |
4617 |
0.10399791688816247 |
No Hit |
| TAAAACATCGCCTACAGAAAAGCGTATGAAAGGAGTGTAAGCTTTGCTCT |
4582 |
0.10320954194965573 |
No Hit |
| GGGATCATAAAGCTGTTGGAAGATGACCTTTTGATAAATGGCAACACTTC |
4519 |
0.10179046706034356 |
No Hit |
| CCAGAATTCCTTCTCTGCTGGAGGCACACTCCCCCAAGGGCACAAAGGAA |
4508 |
0.10154269207967002 |
No Hit |
| ATGTCAGATATAAAGAACTCACCGAACAGCAGCTCCCAGGCGCACTTCCT |
4484 |
0.10100209212183682 |
No Hit |
| CACACTGTCATTAGTTGCAGCATCTCCTGTAGCAATTGGGCTAAATCTGT |
4463 |
0.10052906715873276 |
No Hit |
| TAAGAAATTGCCACACACACAGCCATCCCAGCAACCTTCAGCAACTACCA |
4445 |
0.10012361719035785 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality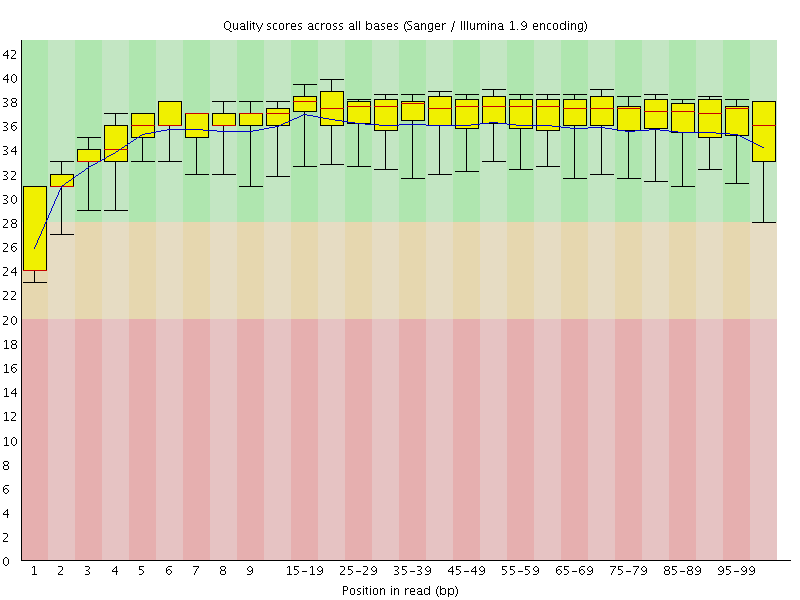
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores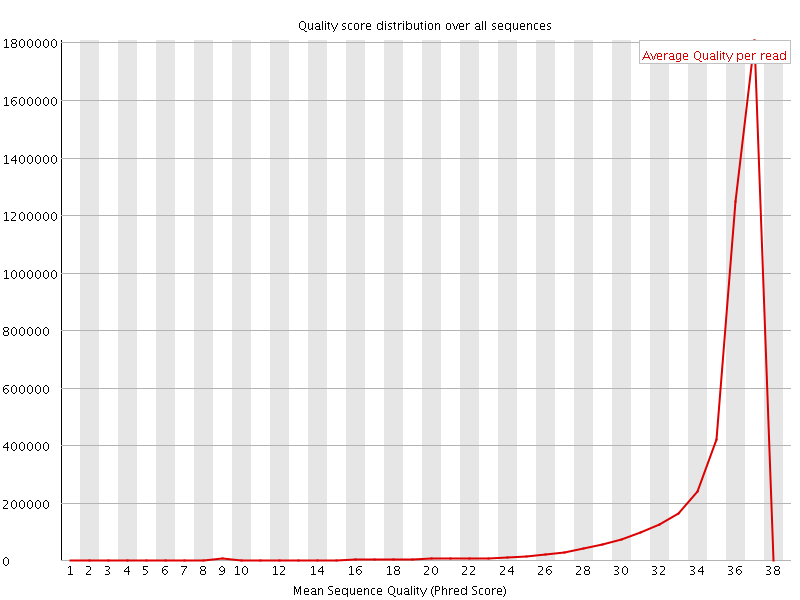
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content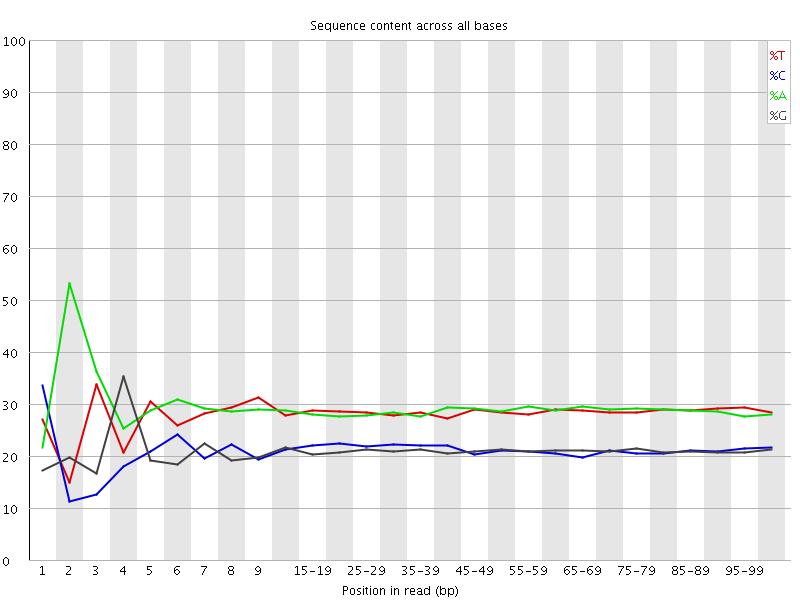
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content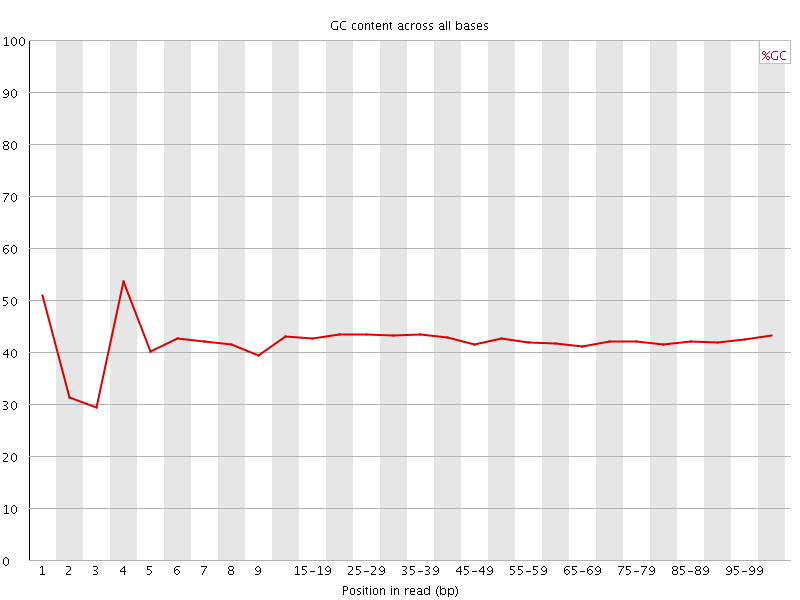
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content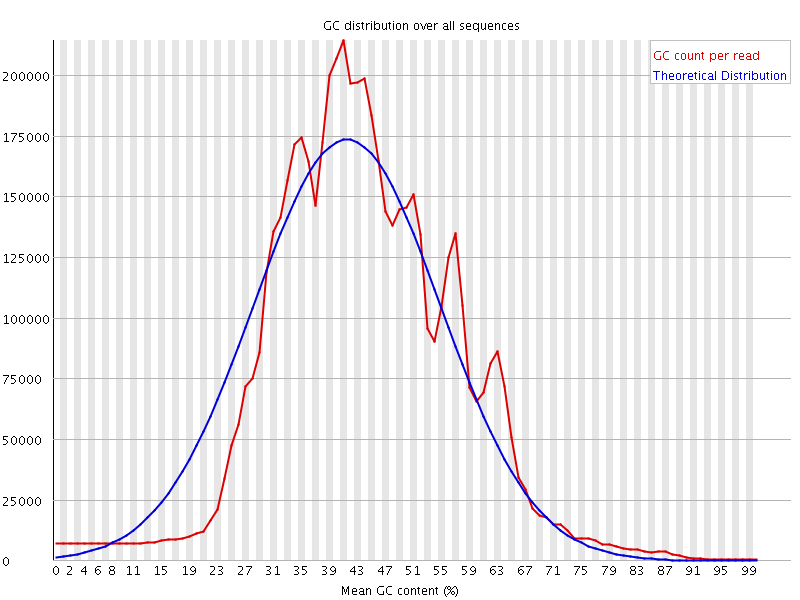
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution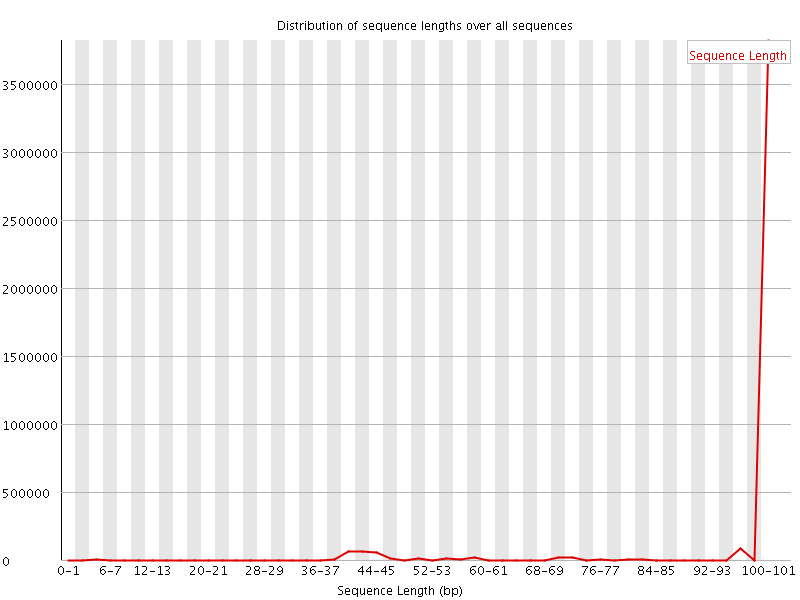
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels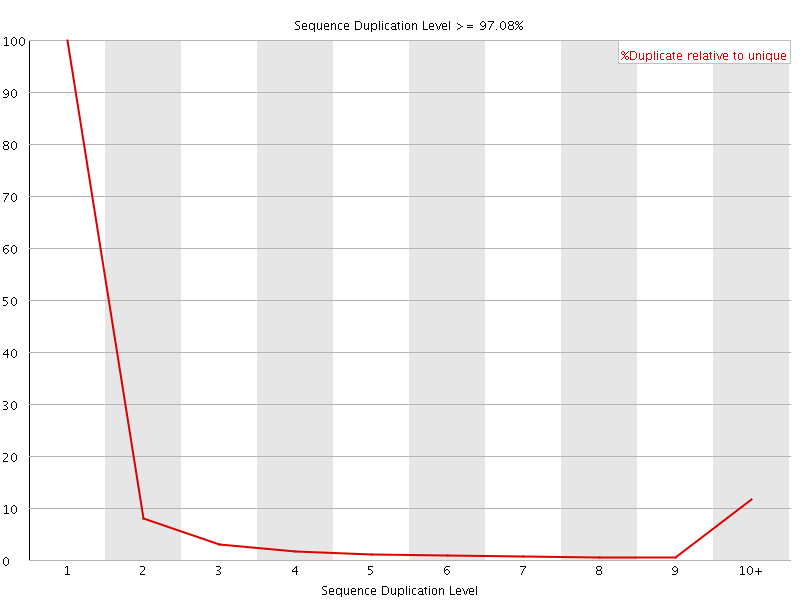
![[WARN]](Icons/warning.png) Overrepresented sequences
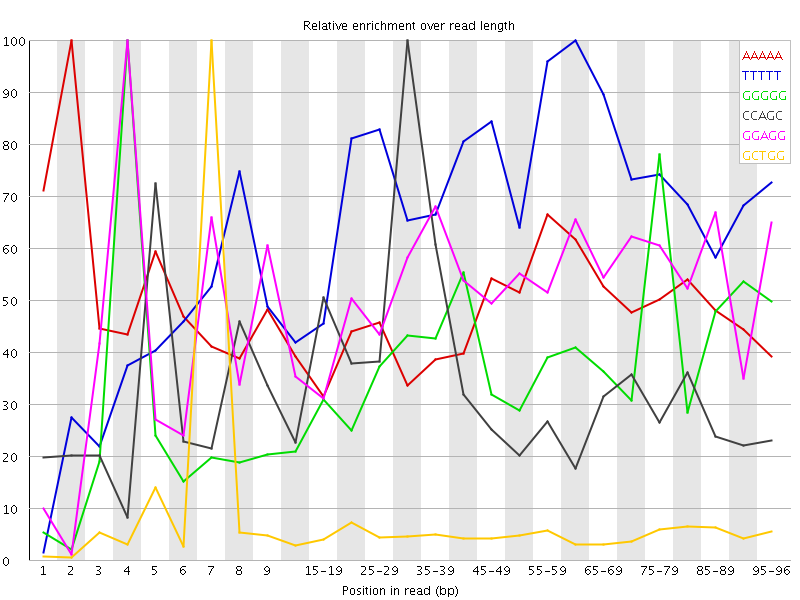
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content