![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 8016-8-55.h.sapiens.aligned.bam |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 12306976 |
| Filtered Sequences | 0 |
| Sequence length | 1-100 |
| %GC | 43 |
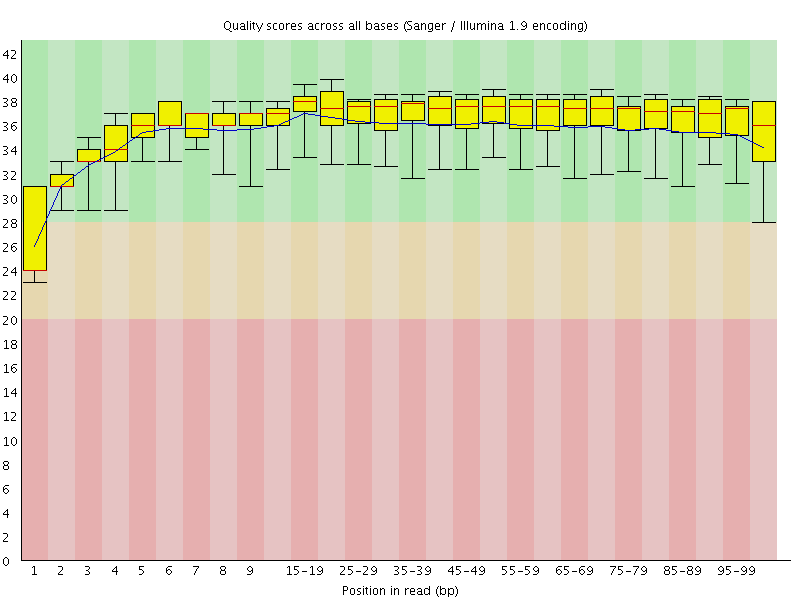
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
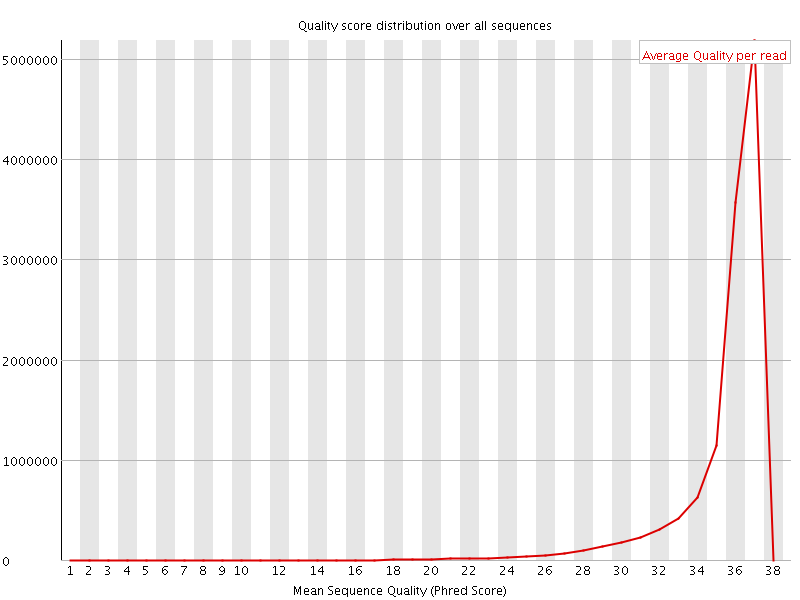
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
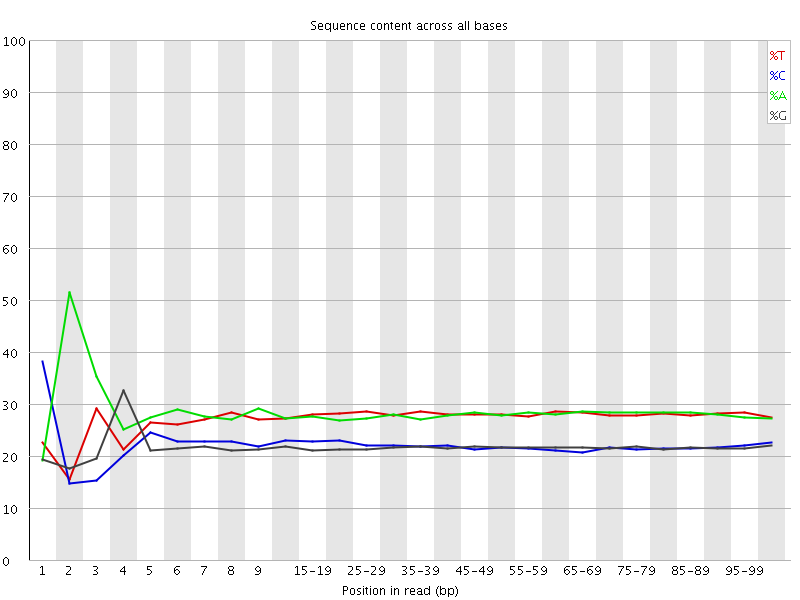
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
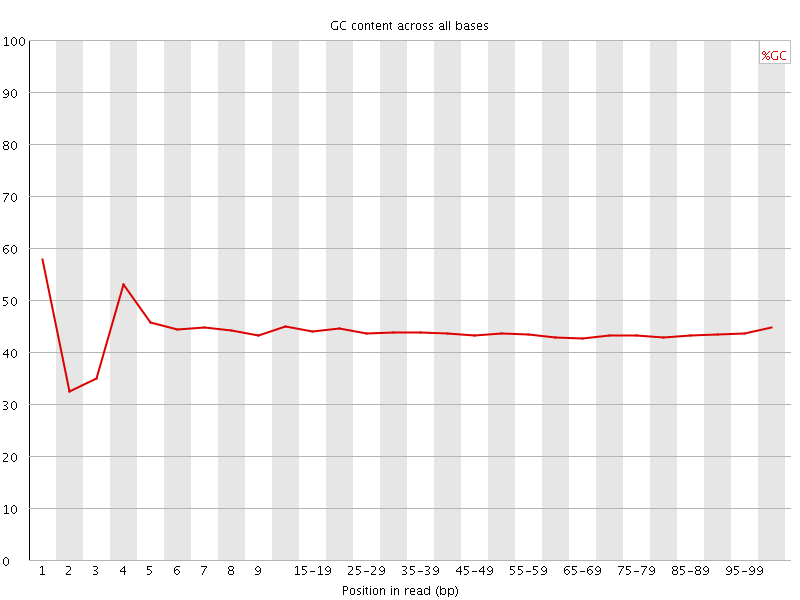
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
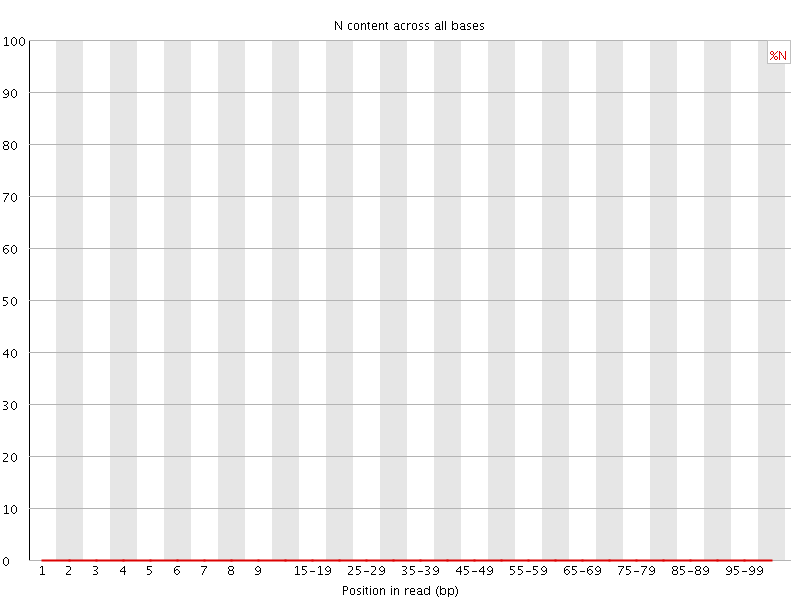
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
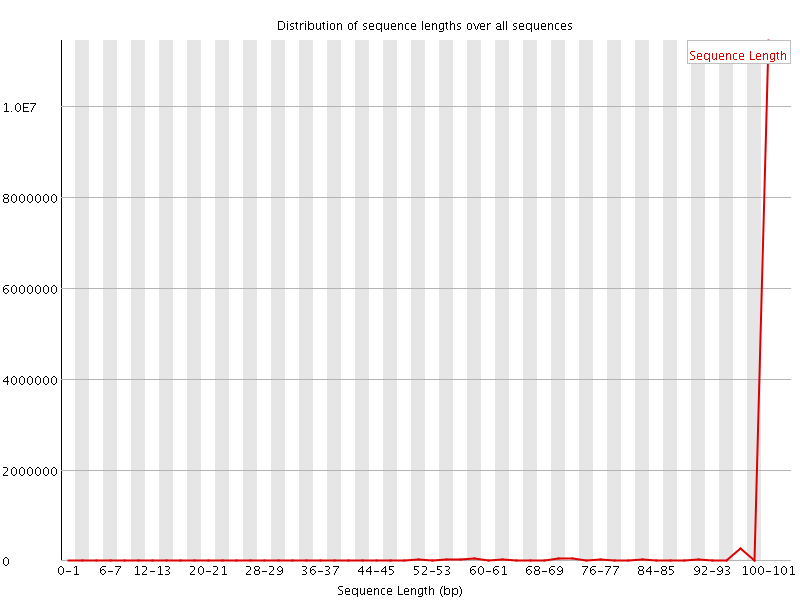
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATGCATCTAAATCTTCTCTGGAGATTATCTCCCTACTGTGTAGGTTAAG | 13293 | 0.10801191129323727 | No Hit |
| CTTGGGGGAGTGTGCCTCCAGCAGAGAAGTAATTCTGGCCTGGCAACCTA | 13225 | 0.1074593791358657 | No Hit |
| GATGGCACACACACACAACACAGGGAGCTGGAGCCTCAGTTTCACGCTGA | 13122 | 0.10662245542690585 | No Hit |
| CATGTAGACTTACCTTTCCTCATAGAGCTATCCTGGTTAATAACAGGCCA | 12732 | 0.10345352099492192 | No Hit |
| ACACTTACACCAAGGAATCTCCCAGGCATTATCAAAGCTACAGGAGACAA | 12674 | 0.10298224356657558 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
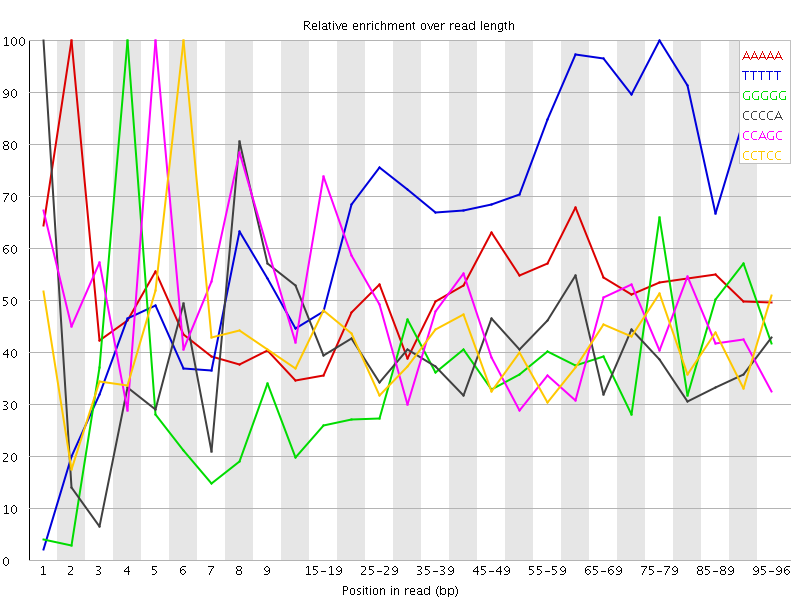
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 6528555 | 3.1129637 | 6.047213 | 2 |
| TTTTT | 6075950 | 3.0730708 | 4.2387924 | 75-79 |
| GGGGG | 1482365 | 2.6290154 | 7.1054316 | 4 |
| CCCCA | 1968995 | 2.5378308 | 6.250884 | 1 |
| CCAGC | 1874605 | 2.450523 | 5.2493744 | 5 |
| CCTCC | 1870075 | 2.4389186 | 5.9475155 | 6 |
| TGGGG | 1707090 | 2.355692 | 9.241428 | 3 |
| CTGCC | 1770815 | 2.3422995 | 5.156597 | 7 |
| GAGGG | 1533480 | 2.0913172 | 5.129274 | 8 |
| TGGCT | 1825260 | 1.932324 | 5.291963 | 3 |
| TGCAG | 1792320 | 1.8752124 | 7.3820763 | 3 |
| GCCCA | 1384060 | 1.8092722 | 5.9888515 | 2 |
| GGCTC | 1311740 | 1.759738 | 7.252563 | 1 |
| TGCCC | 1180735 | 1.5617867 | 8.131325 | 3 |
| GCTCC | 1171730 | 1.5498754 | 6.486392 | 2 |
| GGCCC | 899405 | 1.5289755 | 6.634688 | 1 |
| CATGG | 1441390 | 1.5080523 | 38.332424 | 1 |
| TAAAA | 3069500 | 1.4809653 | 7.8966537 | 1 |
| ATGGA | 1785415 | 1.4568298 | 10.120607 | 2 |
| GCAGT | 1315500 | 1.3763402 | 5.4161787 | 4 |
| ATGTG | 1583490 | 1.3073903 | 8.856091 | 2 |
| TGGGT | 1211820 | 1.3011407 | 5.5415783 | 3 |
| ATGGC | 1223220 | 1.2797923 | 9.633906 | 2 |
| ATGAA | 1985975 | 1.2460837 | 7.8146315 | 2 |
| ATGGG | 1173360 | 1.2450798 | 12.336829 | 2 |
| ATGCC | 1154980 | 1.1914572 | 7.8162017 | 2 |
| CATGT | 1340625 | 1.0913557 | 29.025532 | 1 |
| ATGCT | 1306095 | 1.063246 | 7.791496 | 2 |
| CATGA | 1320245 | 1.062168 | 24.363287 | 1 |
| ATGTT | 1625490 | 1.0442351 | 6.686327 | 2 |
| ATGGT | 1259205 | 1.0396482 | 6.470949 | 2 |
| TAAAG | 1608845 | 1.0094566 | 5.4037213 | 1 |
| CATGC | 940135 | 0.9698269 | 29.70644 | 1 |
| ATGAT | 1517840 | 0.9636509 | 5.0574293 | 2 |
| ATGAC | 1189195 | 0.95673525 | 5.831439 | 2 |
| ATGTA | 1434530 | 0.91075873 | 6.545614 | 2 |
| ATGCA | 1040880 | 0.8374125 | 9.906194 | 2 |
| TAAGA | 1303060 | 0.81759435 | 5.011645 | 1 |
| TAATG | 1208700 | 0.7673831 | 5.3097763 | 1 |
| TAAGT | 1171185 | 0.7435655 | 6.9323664 | 1 |
| TAAAC | 1100775 | 0.6809905 | 5.2067094 | 1 |
| TAAGG | 828045 | 0.6756528 | 5.999074 | 1 |
| TGCCG | 385105 | 0.51662976 | 7.5480857 | 5 |