The Select Menu
The items in this menu are used to modify the current selection (see the section called The Selection in Chapter 1). Artemis A short summary of the current selection is shown at the top of the main window (see 2 for details)..
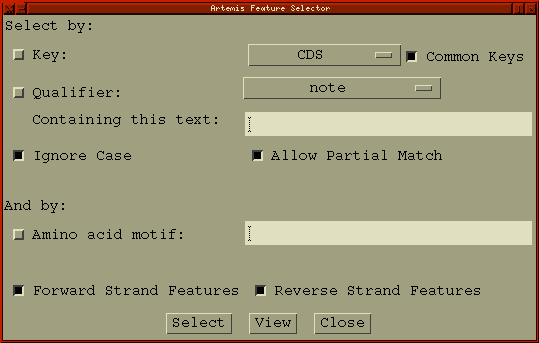
Feature Selector ...
Open a new Feature Selector window. This window allows the user to choose which features to select or view based on feature keys (see the section called EMBL/Genbank Feature Keys in Chapter 1), qualifier values (see the section called EMBL/Genbank Feature Qualifiers in Chapter 1) and amino acid motifs.
The Select button will set the selection to the contain those features that match the given key, qualifier and amino acid motif combination.
The View button will create a new feature list (see the section called The Feature List) containing only those features that match the given key, qualifier and amino acid motif combination.
All
Reset the selection so that nothing is selected then select all the features in the active entries. [shortcut key: A]
All Bases
Reset the selection so that nothing is selected then select all the bases in the sequence.
Select All Features in Non-matching Regions
Select all features that have no corresponding match in ACT. This is used to higlight regions that are different between sets of sequence. It will only take into account matches that have not been filtered out using the score, identity or length cut-off.
None
Clear the selection so that nothing is selected. [shortcut key: N]
By Key
Ask the user for a feature key, reset the selection so that nothing is selected, then select all the features with the key given by the user.
CDS Features
Reset the selection so that nothing is selected, then select all the CDS features that do not have a /pseudo qualifier.
Same Key
Select all the features that have the same key as any of the currently selected features.
Open Reading Frame
Extend the current selection of bases to cover complete open reading frames. Selecting a single base or codon and then choosing this menu item has a similar effect to double clicking the middle button on a base or residue (see the section called Changing the Selection from a View Window for details).
Features Overlapping Selection
Select those (and only those) features that overlap the currently selected range of bases or any of the currently selected features. The current selection will be discarded.
Base Range ...
Ask the user for a range of bases, then select those bases. The range should look something like this: 100-200, complement(100..200), 100.200 or 100..200. If the first number is larger than the second the bases will be selected on the forward strand, otherwise they will be selected on the reverse strand (unless there is a complement around the range, in which case the sense is reversed).
Feature AA Range ...
Ask the user for a range of amino acids in the selected feature and select those bases. The range should look something like this: 100-200, or 100..200.
Toggle Selection
Invert the selection - after choosing this menu item the selection will contain only those features that were not in the selection beforehand.